Deleterious heteroplasmic mitochondrial mutations are associated with an increased risk of overall and cancer-specific mortality
- PMID: 37777527
- PMCID: PMC10542802
- DOI: 10.1038/s41467-023-41785-7
Deleterious heteroplasmic mitochondrial mutations are associated with an increased risk of overall and cancer-specific mortality
Abstract
Mitochondria carry their own circular genome and disruption of the mitochondrial genome is associated with various aging-related diseases. Unlike the nuclear genome, mitochondrial DNA (mtDNA) can be present at 1000 s to 10,000 s copies in somatic cells and variants may exist in a state of heteroplasmy, where only a fraction of the DNA molecules harbors a particular variant. We quantify mtDNA heteroplasmy in 194,871 participants in the UK Biobank and find that heteroplasmy is associated with a 1.5-fold increased risk of all-cause mortality. Additionally, we functionally characterize mtDNA single nucleotide variants (SNVs) using a constraint-based score, mitochondrial local constraint score sum (MSS) and find it associated with all-cause mortality, and with the prevalence and incidence of cancer and cancer-related mortality, particularly leukemia. These results indicate that mitochondria may have a functional role in certain cancers, and mitochondrial heteroplasmic SNVs may serve as a prognostic marker for cancer, especially for leukemia.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures
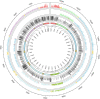

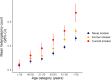
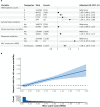


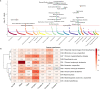
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL092577/HL/NHLBI NIH HHS/United States
- 75N92020D00001/HL/NHLBI NIH HHS/United States
- U01 HL120393/HL/NHLBI NIH HHS/United States
- N01 HC095167/HL/NHLBI NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- R01 HL156144/HL/NHLBI NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- HHSN268201800001C/HL/NHLBI NIH HHS/United States
- R01 HL144569/HL/NHLBI NIH HHS/United States
- UL1 TR000040/TR/NCATS NIH HHS/United States
- N01 HC095160/HL/NHLBI NIH HHS/United States
- 75N92020D00002/HL/NHLBI NIH HHS/United States
- HHSN268201500003C/HL/NHLBI NIH HHS/United States
- N01 HC095161/HL/NHLBI NIH HHS/United States
- 75N92020D00005/HL/NHLBI NIH HHS/United States
- N01 HC095168/HL/NHLBI NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- R01 HL131136/HL/NHLBI NIH HHS/United States
- UL1 TR001079/TR/NCATS NIH HHS/United States
- N01 HC095169/HL/NHLBI NIH HHS/United States
- N01 HC095159/HL/NHLBI NIH HHS/United States
- 75N92020D00003/HL/NHLBI NIH HHS/United States
- HHSN268201500015C/HL/NHLBI NIH HHS/United States
- K24 HL148521/HL/NHLBI NIH HHS/United States
- 75N92020D00004/HL/NHLBI NIH HHS/United States
- UL1 TR001420/TR/NCATS NIH HHS/United States
- R01 AG059727/AG/NIA NIH HHS/United States
- N01 HC095163/HL/NHLBI NIH HHS/United States
- 75N92020D00007/HL/NHLBI NIH HHS/United States
- HHSN268201500014C/HL/NHLBI NIH HHS/United States
- HHSN268201500003I/HL/NHLBI NIH HHS/United States
- N01 HC095166/HL/NHLBI NIH HHS/United States
- UM1 HG008898/HG/NHGRI NIH HHS/United States
- 75N92020D00006/HL/NHLBI NIH HHS/United States
- R01 HL117626/HL/NHLBI NIH HHS/United States
- N01 HC095162/HL/NHLBI NIH HHS/United States
- UL1 TR001881/TR/NCATS NIH HHS/United States
- U24 HG008956/HG/NHGRI NIH HHS/United States
- N01 HC095165/HL/NHLBI NIH HHS/United States
- N01 HC095164/HL/NHLBI NIH HHS/United States

