Selection and enrichment of microbial species with an increased lignocellulolytic phenotype from a native soil microbiome by activity-based probing
- PMID: 37777628
- PMCID: PMC10542759
- DOI: 10.1038/s43705-023-00305-w
Selection and enrichment of microbial species with an increased lignocellulolytic phenotype from a native soil microbiome by activity-based probing
Abstract
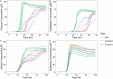
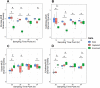
Multi-omic analyses can provide information on the potential for activity within a microbial community but often lack specificity to link functions to cell, primarily offer potential for function or rely on annotated databases. Functional assays are necessary for understanding in situ microbial activity to better describe and improve microbiome biology. Targeting enzyme activity through activity-based protein profiling enhances the accuracy of functional studies. Here, we introduce a pipeline of coupling activity-based probing with fluorescence-activated cell sorting, culturing, and downstream activity assays to isolate and examine viable populations of cells expressing a function of interest. We applied our approach to a soil microbiome using two activity-based probes to enrich for communities with elevated activity for lignocellulose-degradation phenotypes as determined by four fluorogenic kinetic assays. Our approach efficiently separated and identified microbial members with heightened activity for glycosyl hydrolases, and by expanding this workflow to various probes for other function, this process can be applied to unique phenotype targets of interest.
© 2023. ISME Publications B.V.
Conflict of interest statement
ATW is a member of the scientific advisory board of Enzymetrics Biosciences.
Figures



References
-
- Biessy L, Pearman JK, Waters S, Vandergoes MJ, Wood SA. Metagenomic insights to the functional potential of sediment microbial communities in freshwater lakes. Metabarcoding Metagenom. 2022;6:59–74. doi: 10.3897/mbmg.6.79265. - DOI

