Influence of variant-specific mutations, temperature and pH on conformations of a large set of SARS-CoV-2 spike trimer vaccine antigen candidates
- PMID: 37779126
- PMCID: PMC10543594
- DOI: 10.1038/s41598-023-43661-2
Influence of variant-specific mutations, temperature and pH on conformations of a large set of SARS-CoV-2 spike trimer vaccine antigen candidates
Abstract
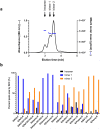
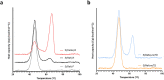
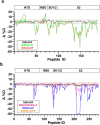
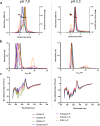
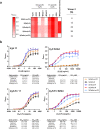
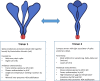
SARS-CoV-2 subunit vaccines continue to be the focus of intense clinical development worldwide. Protein antigens in these vaccines most commonly consist of the spike ectodomain fused to a heterologous trimerization sequence, designed to mimic the compact, prefusion conformation of the spike on the virus surface. Since 2020, we have produced dozens of such constructs in CHO cells, consisting of spike variants with different mutations fused to different trimerization sequences. This set of constructs displayed notable conformational heterogeneity, with two distinct trimer species consistently detected by analytical size exclusion chromatography. A recent report showed that spike ectodomain fusion constructs can adopt an alternative trimer conformation consisting of loosely associated ectodomain protomers. Here, we applied multiple biophysical and immunological techniques to demonstrate that this alternative conformation is formed to a significant extent by several SARS-CoV-2 variant spike proteins. We have also examined the influence of temperature and pH, which can induce inter-conversion of the two forms. The substantial structural differences between these trimer types may impact their performance as vaccine antigens.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Gilbert PB, et al. A COVID-19 milestone attained—A correlate of protection for vaccines. N. Engl. J. Med. 2022;387:2203–2206. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

