Archaeome structure and function of the intestinal tract in healthy and H1N1 infected swine
- PMID: 37779690
- PMCID: PMC10534045
- DOI: 10.3389/fmicb.2023.1250140
Archaeome structure and function of the intestinal tract in healthy and H1N1 infected swine
Abstract
Background: Methanogenic archaea represent a less investigated and likely underestimated part of the intestinal tract microbiome in swine.
Aims/methods: This study aims to elucidate the archaeome structure and function in the porcine intestinal tract of healthy and H1N1 infected swine. We performed multi-omics analysis consisting of 16S rRNA gene profiling, metatranscriptomics and metaproteomics.
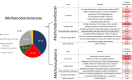
Results and discussion: We observed a significant increase from 0.48 to 4.50% of archaea in the intestinal tract microbiome along the ileum and colon, dominated by genera Methanobrevibacter and Methanosphaera. Furthermore, in feces of naïve and H1N1 infected swine, we observed significant but minor differences in the occurrence of archaeal phylotypes over the course of an infection experiment. Metatranscriptomic analysis of archaeal mRNAs revealed the major methanogenesis pathways of Methanobrevibacter and Methanosphaera to be hydrogenotrophic and methyl-reducing, respectively. Metaproteomics of archaeal peptides indicated some effects of the H1N1 infection on central metabolism of the gut archaea.
Conclusions/take home message: Finally, this study provides the first multi-omics analysis and high-resolution insights into the structure and function of the porcine intestinal tract archaeome during a non-lethal Influenza A virus infection of the respiratory tract, demonstrating significant alterations in archaeal community composition and central metabolic functions.
Keywords: 16S rRNA gene sequencing; intestinal tract microbiome; metaproteomics; metatranscriptomics; methanogenesis; methanogenic archaea.
Copyright © 2023 Meene, Gierse, Schwaiger, Karte, Schröder, Höper, Wang, Groß, Wünsche, Mücke, Kreikemeyer, Beer, Becher, Mettenleiter, Riedel and Urich.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources

