Single-Nucleotide Resolution Mapping of N6-Methyladenine in Genomic DNA
- PMID: 37780356
- PMCID: PMC10540296
- DOI: 10.1021/acscentsci.3c00481
Single-Nucleotide Resolution Mapping of N6-Methyladenine in Genomic DNA
Abstract
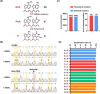
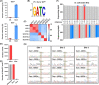
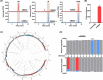
N6-Methyladenine (6mA) is a naturally occurring DNA modification in both prokaryotes and eukaryotes. Herein, we developed a deaminase-mediated sequencing (DM-seq) method for genome-wide mapping of 6mA at single-nucleotide resolution. The method capitalizes on the selective deamination of adenine, but not 6mA, in DNA mediated by an evolved adenine deaminase, ABE8e. By employing this method, we achieved genome-wide mapping of 6mA in Escherichia coli and in mammalian mitochondrial DNA (mtDNA) at single-nucleotide resolution. We found that the 6mA sites are mainly located in the GATC motif in the E. coli genome. We also identified 17 6mA sites in mtDNA of HepG2 cells, where all of the 6mA sites are distributed in the heavy strand of mtDNA. We envision that DM-seq will be a valuable tool for uncovering new functions of 6mA in DNA and for exploring its potential roles in mitochondria-related human diseases.
© 2023 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures