Evidence supportive of a bacterial component in the etiology for Alzheimer's disease and for a temporal-spatial development of a pathogenic microbiome in the brain
- PMID: 37780846
- PMCID: PMC10534976
- DOI: 10.3389/fcimb.2023.1123228
Evidence supportive of a bacterial component in the etiology for Alzheimer's disease and for a temporal-spatial development of a pathogenic microbiome in the brain
Abstract
Background: Over the last few decades, a growing body of evidence has suggested a role for various infectious agents in Alzheimer's disease (AD) pathogenesis. Despite diverse pathogens (virus, bacteria, fungi) being detected in AD subjects' brains, research has focused on individual pathogens and only a few studies investigated the hypothesis of a bacterial brain microbiome. We profiled the bacterial communities present in non-demented controls and AD subjects' brains.
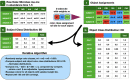
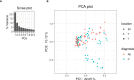
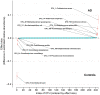
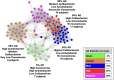
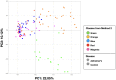
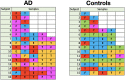
Results: We obtained postmortem samples from the brains of 32 individual subjects, comprising 16 AD and 16 control age-matched subjects with a total of 130 samples from the frontal and temporal lobes and the entorhinal cortex. We used full-length 16S rRNA gene amplification with Pacific Biosciences sequencing technology to identify bacteria. We detected bacteria in the brains of both cohorts with the principal bacteria comprising Cutibacterium acnes (formerly Propionibacterium acnes) and two species each of Acinetobacter and Comamonas genera. We used a hierarchical Bayesian method to detect differences in relative abundance among AD and control groups. Because of large abundance variances, we also employed a new analysis approach based on the Latent Dirichlet Allocation algorithm, used in computational linguistics. This allowed us to identify five sample classes, each revealing a different microbiota. Assuming that samples represented infections that began at different times, we ordered these classes in time, finding that the last class exclusively explained the existence or non-existence of AD.
Conclusions: The AD-related pathogenicity of the brain microbiome seems to be based on a complex polymicrobial dynamic. The time ordering revealed a rise and fall of the abundance of C. acnes with pathogenicity occurring for an off-peak abundance level in association with at least one other bacterium from a set of genera that included Methylobacterium, Bacillus, Caulobacter, Delftia, and Variovorax. C. acnes may also be involved with outcompeting the Comamonas species, which were strongly associated with non-demented brain microbiota, whose early destruction could be the first stage of disease. Our results are also consistent with a leaky blood-brain barrier or lymphatic network that allows bacteria, viruses, fungi, or other pathogens to enter the brain.
Keywords: 16s sequencing; Alzheimer’s disease; Bayesian; Cutibacterium; blood brain barrier; glymphatic network; latent dirichlet allocation; microbiome.
Copyright © 2023 Moné, Earl, Król, Ahmed, Sen, Ehrlich and Lapides.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








Similar articles
-
Much ado about nothing? Off-target amplification can lead to false-positive bacterial brain microbiome detection in healthy and Parkinson's disease individuals.Microbiome. 2021 Mar 26;9(1):75. doi: 10.1186/s40168-021-01012-1. Microbiome. 2021. PMID: 33771222 Free PMC article.
-
Association of Systemic Antibiotic Treatment of Acne With Skin Microbiota Characteristics.JAMA Dermatol. 2019 Apr 1;155(4):425-434. doi: 10.1001/jamadermatol.2018.5221. JAMA Dermatol. 2019. PMID: 30758497 Free PMC article.
-
Analysis of Salivary Microbiome in Patients with Alzheimer's Disease.J Alzheimers Dis. 2019;72(2):633-640. doi: 10.3233/JAD-190587. J Alzheimers Dis. 2019. PMID: 31594229
-
The increasing importance of the gut microbiome in acne vulgaris.Folia Microbiol (Praha). 2022 Dec;67(6):825-835. doi: 10.1007/s12223-022-00982-5. Epub 2022 Jun 16. Folia Microbiol (Praha). 2022. PMID: 35711021 Review.
-
Recent advances in understanding Propionibacterium acnes ( Cutibacterium acnes) in acne.F1000Res. 2018 Dec 19;7:F1000 Faculty Rev-1953. doi: 10.12688/f1000research.15659.1. eCollection 2018. F1000Res. 2018. PMID: 30613388 Free PMC article. Review.
Cited by
-
Detection of fungal sequences in human brain: rDNA locus amplification and deep sequencing.Sci Rep. 2024 Dec 30;14(1):31790. doi: 10.1038/s41598-024-82840-7. Sci Rep. 2024. PMID: 39738312 Free PMC article.
-
Establishment of a consensus protocol to explore the brain pathobiome in patients with mild cognitive impairment and Alzheimer's disease: Research outline and call for collaboration.Alzheimers Dement. 2023 Nov;19(11):5209-5231. doi: 10.1002/alz.13076. Epub 2023 Jun 7. Alzheimers Dement. 2023. PMID: 37283269 Free PMC article. Review.
-
Regional Differences in Microbial Infiltration of Brain Tissue from Alzheimer's Disease Patients and Control Individuals.Brain Sci. 2024 Jul 3;14(7):677. doi: 10.3390/brainsci14070677. Brain Sci. 2024. PMID: 39061418 Free PMC article.
-
The blood-brain barrier: a help and a hindrance.Brain. 2025 Jul 7;148(7):2262-2282. doi: 10.1093/brain/awaf068. Brain. 2025. PMID: 39969549 Free PMC article. Review.
-
m5C RNA methylation: a potential mechanism for infectious Alzheimer's disease.Front Cell Dev Biol. 2024 Aug 8;12:1440143. doi: 10.3389/fcell.2024.1440143. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39175875 Free PMC article. Review.
References
-
- Al-Atrache Z., Lopez D. B., Hingley S. T., Appelt D. M. (2019). Astrocytes infected with Chlamydia pneumoniae demonstrate altered expression and activity of secretases involved in the generation of ?-amyloid found in Alzheimer disease. BMC Neurosci. 20, 6. doi: 10.1186/s12868-019-0489-5 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

