Diagnostic efficiency of metagenomic next-generation sequencing for suspected infection in allogeneic hematopoietic stem cell transplantation recipients
- PMID: 37780852
- PMCID: PMC10533937
- DOI: 10.3389/fcimb.2023.1251509
Diagnostic efficiency of metagenomic next-generation sequencing for suspected infection in allogeneic hematopoietic stem cell transplantation recipients
Abstract
Introduction: Immunosuppression predisposes allogeneic hematopoietic stem cell transplantation (allo-HSCT) recipients to infection. Prompt and accurate identification of pathogens is crucial to optimize treatment strategies. This multi-center retrospective study aimed to assess the ability of metagenomic next-generation sequencing (mNGS) to detect causative pathogens in febrile allo-HSCT recipients and examined its concordance with conventional microbiological tests (CMT).
Methods: We performed mNGS and CMT on samples obtained from 153 patients with suspected infection during allo-HSCT. Patients were grouped based on their neutropenic status at the time of sampling.
Results: The mNGS test was more sensitive than CMT (81.1% vs. 53.6%, P<0.001) for diagnosing clinically suspected infection, especially in the non-neutropenia cohort. mNGS could detect fungi and viruses better than bacteria, with a higher sensitivity than CMT. Immune events were diagnosed in 57.4% (35/61) of the febrile events with negative mNGS results, and 33.5% (48/143) with negative CMT results (P=0.002). The treatment success rate of the targeted anti-infection strategy was significantly higher when based on mNGS than on empirical antibiotics (85% vs. 56.5%, P=0.004).
Conclusion: The mNGS test is superior to CMT for identifying clinically relevant pathogens, and provides valuable information for anti-infection strategies in allo-HSCT recipients. Additionally, attention should be paid to immune events in patients with negative mNGS results.
Keywords: clinical infection; conventional microbiological tests; diagnostic efficiency; immune events; metagenomic next-generation sequencing.
Copyright © 2023 Huang, Zhao, Jiang, Han, Pan, Zhang, Wang, Chen, Li, Zhao and Hu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
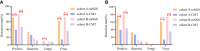


Similar articles
-
Application of plasma metagenomic next-generation sequencing improves prognosis in hematology patients with neutropenia or hematopoietic stem cell transplantation for infection.Front Cell Infect Microbiol. 2024 Feb 2;14:1338307. doi: 10.3389/fcimb.2024.1338307. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38371300 Free PMC article.
-
Metagenomic Next-Generation Sequencing in the Diagnosis of Pulmonary Infections after Allogeneic Hematopoietic Stem Cell Transplantation.Transplant Cell Ther. 2024 Nov;30(11):1110.e1-1110.e10. doi: 10.1016/j.jtct.2024.08.014. Epub 2024 Aug 27. Transplant Cell Ther. 2024. PMID: 39197493
-
Diagnostic performance and clinical impacts of metagenomic sequencing after allogeneic hematopoietic stem cell transplantation.J Microbiol Immunol Infect. 2024 Feb;57(1):11-19. doi: 10.1016/j.jmii.2023.11.002. Epub 2023 Nov 28. J Microbiol Immunol Infect. 2024. PMID: 38065767
-
Clinical and diagnostic values of metagenomic next-generation sequencing for infection in hematology patients: a systematic review and meta-analysis.BMC Infect Dis. 2024 Feb 7;24(1):167. doi: 10.1186/s12879-024-09073-x. BMC Infect Dis. 2024. PMID: 38326763 Free PMC article.
-
The role of metagenomic next-generation sequencing in diagnosing and managing post-kidney transplantation infections.Front Cell Infect Microbiol. 2025 Jan 7;14:1473068. doi: 10.3389/fcimb.2024.1473068. eCollection 2024. Front Cell Infect Microbiol. 2025. PMID: 39839264 Free PMC article. Review.
Cited by
-
Metagenomic versus targeted next-generation sequencing for detection of microorganisms in bronchoalveolar lavage fluid among renal transplantation recipients.Front Immunol. 2024 Aug 26;15:1443057. doi: 10.3389/fimmu.2024.1443057. eCollection 2024. Front Immunol. 2024. PMID: 39253087 Free PMC article.
-
Targeted Next-Generation Sequencing in Pneumonia: Applications in the Detection of Responsible Pathogens, Antimicrobial Resistance, and Virulence.Infect Drug Resist. 2025 Jan 22;18:407-418. doi: 10.2147/IDR.S504392. eCollection 2025. Infect Drug Resist. 2025. PMID: 39872133 Free PMC article.
-
Outcomes of acute myeloid leukemia with KMT2A (MLL) rearrangement: a multicenter study of TROPHY group.Blood Cancer J. 2025 May 2;15(1):84. doi: 10.1038/s41408-025-01293-x. Blood Cancer J. 2025. PMID: 40316511 Free PMC article.
References
-
- Busca A., Cinatti N., Gill J., Passera R., Dellacasa C. M., Giaccone L., et al. . (2021). Management of invasive fungal infections in patients undergoing allogeneic hematopoietic stem cell transplantation: the Turin experience. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.805514 - DOI - PMC - PubMed
-
- Chang Y. J., Wang Y., Xu L. P., Zhang X. H., Chen H., Chen Y. H., et al. . (2020). Haploidentical donor is preferred over matched sibling donor for pre-transplantation MRD positive ALL: a phase 3 genetically randomized study. J. Hematol. Oncol. 13 (1), 27. doi: 10.1186/s13045-020-00860-y - DOI - PMC - PubMed
-
- Chemaly R. F., El Haddad L., Winston D. J., Rowley S. D., Mulane K. M., Chandrasekar P., et al. . (2020). Cytomegalovirus (CMV) cell-mediated immunity and CMV infection after allogeneic hematopoietic cell transplantation: the REACT study. Clin. Infect. Dis. 71 (9), 2365–2374. doi: 10.1093/cid/ciz1210 - DOI - PMC - PubMed
-
- Chen L., Wang Y., Zhang T., Li Y., Meng T., Liu L., et al. . (2018). Utility of posaconazole therapeutic drug monitoring and assessment of plasma concentration threshold for effective prophylaxis of invasive fungal infections: a meta-analysis with trial sequential analysis. BMC Infect. Dis. 18 (1), 155. doi: 10.1186/s12879-018-3055-3 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

