Glycine Decarboxylase (GLDC) Plays a Crucial Role in Regulating Energy Metabolism, Invasion, Metastasis and Immune Escape for Prostate Cancer
- PMID: 37781511
- PMCID: PMC10539704
- DOI: 10.7150/ijbs.85893
Glycine Decarboxylase (GLDC) Plays a Crucial Role in Regulating Energy Metabolism, Invasion, Metastasis and Immune Escape for Prostate Cancer
Abstract
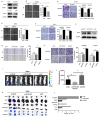
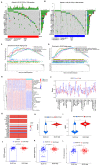
Glycine decarboxylase (GLDC) is one of the core enzymes for glycine metabolism, and its biological roles in prostate cancer (PCa) are unclear. First, we found that GLDC plays a central role in glycolysis in 540 TCGA PCa patients. Subsequently, a metabolomic microarray showed that GLDC enhanced aerobic glycolysis in PCa cells, and GLDC and its enzyme activity enhanced glucose uptake, lactate production and lactate dehydrogenase (LDH) activity in PCa cells. Next, we found that GLDC was highly expressed in PCa, was directly regulated by hypoxia-inducible factor (HIF1-α) and regulated downstream LDHA expression. In addition, GLDC and its enzyme activity showed a strong ability to promote the migration and invasion of PCa both in vivo and in vitro. Furthermore, we found that the GLDC-high group had a higher TP53 mutation frequency, lower CD8+ T-cell infiltration, higher immune checkpoint expression, and higher immune exclusion scores than the GLDC-low group. Finally, the GLDC-based prognostic risk model by applying LASSO Cox regression also showed good predictive power for the clinical characteristics and survival in PCa patients. This evidence indicates that GLDC plays crucial roles in glycolytic metabolism, invasion and metastasis, and immune escape in PCa, and it is a potential therapeutic target for prostate cancer.
Keywords: Glycine decarboxylase (GLDC); Glycolytic metabolism; Immune escape; Metastasis; Prostate cancer.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures






References
-
- Bray F, Ferlay J, Soerjomataram I. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. - PubMed
-
- Psutka SP, Frank I, Karnes RJ. Risk Stratification in Hormone-sensitive Metastatic Prostate Cancer: More Questions than Answers. Eur Urol. 2015;68:205–206. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

