This is a preprint.
A transcriptionally active lipid vesicle encloses the injected Chimalliviridae genome in early infection
- PMID: 37781618
- PMCID: PMC10541120
- DOI: 10.1101/2023.09.20.558163
A transcriptionally active lipid vesicle encloses the injected Chimalliviridae genome in early infection
Abstract
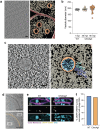
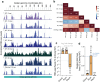
Many eukaryotic viruses require membrane-bound compartments for replication, but no such organelles are known to be formed by prokaryotic viruses1-3. Bacteriophages of the Chimalliviridae family sequester their genomes within a phage-generated organelle, the phage nucleus, which is enclosed by a lattice of the viral protein ChmA4-10. Previously, we observed lipid membrane-bound vesicles in cells infected by Chimalliviridae, but due to the paucity of genetics tools for these viruses it was unknown if these vesicles represented unproductive, abortive infections or a bona fide stage in the phage life cycle. Using the recently-developed dRfxCas13d-based knockdown system CRISPRi-ART11 in combination with fluorescence microscopy and cryo-electron tomography, we show that inhibiting phage nucleus formation arrests infections at an early stage in which the injected phage genome is enclosed within a membrane-bound early phage infection (EPI) vesicle. We demonstrate that early phage genes are transcribed by the virion-associated RNA polymerase from the genome within the compartment, making the EPI vesicle the first known example of a lipid membrane-bound organelle that separates transcription from translation in prokaryotes. Further, we show that the phage nucleus is essential for the phage life cycle, with genome replication only beginning after the injected DNA is transferred from the EPI vesicle to the newly assembled phage nucleus. Our results show that Chimalliviridae require two sophisticated subcellular compartments of distinct compositions and functions that facilitate successive stages of the viral life cycle.
Conflict of interest statement
Competing interest statement K.P. and J.P. have an equity interest in Linnaeus Bioscience Incorporated and receive income. The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its conflict-of-interest policies. The Regents of the University of California have patents issued and pending for CRISPR technologies on which J.A.D., B.F.C., B.A.A., D.F.S., E.C., J.P., K.P., E.G.A. and J.L. are inventors. J.A.D. is a cofounder of Caribou Biosciences, Editas Medicine, Scribe Therapeutics, Intellia Therapeutics, and Mammoth Biosciences. J.A.D. is a scientific advisory board member of Vertex, Caribou Biosciences, Intellia Therapeutics, Scribe Therapeutics, Mammoth Biosciences, Algen Biotechnologies, Felix Biosciences, The Column Group, and Inari. J.A.D. is Chief Science Advisor to Sixth Street, a Director at Johnson & Johnson, Altos and Tempus, and has research projects sponsored by Apple Tree Partners and Roche. D.F.S. is a cofounder and scientific advisory board member of Scribe Therapeutics. All other authors declare no competing interests.
Figures

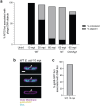











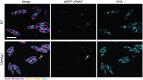

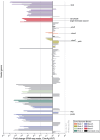
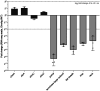






References
-
- Taxonomy Browser. International Committee on Taxonomy of Viruses (ICTV) https://ictv.global/taxonomy.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
