Identifying potential biomarkers of idiopathic pulmonary fibrosis through machine learning analysis
- PMID: 37783761
- PMCID: PMC10545744
- DOI: 10.1038/s41598-023-43834-z
Identifying potential biomarkers of idiopathic pulmonary fibrosis through machine learning analysis
Abstract
Idiopathic pulmonary fibrosis (IPF) is the most common and serious type of idiopathic interstitial pneumonia, characterized by chronic, progressive, and low survival rates, while unknown disease etiology. Until recently, patients with idiopathic pulmonary fibrosis have a poor prognosis, high mortality, and limited treatment options, due to the lack of effective early diagnostic and prognostic tools. Therefore, we aimed to identify biomarkers for idiopathic pulmonary fibrosis based on multiple machine-learning approaches and to evaluate the role of immune infiltration in the disease. The gene expression profile and its corresponding clinical data of idiopathic pulmonary fibrosis patients were downloaded from Gene Expression Omnibus (GEO) database. Next, the differentially expressed genes (DEGs) with the threshold of FDR < 0.05 and |log2 foldchange (FC)| > 0.585 were analyzed via R package "DESeq2" and GO enrichment and KEGG pathways were run in R software. Then, least absolute shrinkage and selection operator (LASSO) logistic regression, support vector machine-recursive feature elimination (SVM-RFE) and random forest (RF) algorithms were combined to screen the key potential biomarkers of idiopathic pulmonary fibrosis. The diagnostic performance of these biomarkers was evaluated through receiver operating characteristic (ROC) curves. Moreover, the CIBERSORT algorithm was employed to assess the infiltration of immune cells and the relationship between the infiltrating immune cells and the biomarkers. Finally, we sought to understand the potential pathogenic role of the biomarker (SLAIN1) in idiopathic pulmonary fibrosis using a mouse model and cellular model. A total of 3658 differentially expressed genes of idiopathic pulmonary fibrosis were identified, including 2359 upregulated genes and 1299 downregulated genes. FHL2, HPCAL1, RNF182, and SLAIN1 were identified as biomarkers of idiopathic pulmonary fibrosis using LASSO logistic regression, RF, and SVM-RFE algorithms. The ROC curves confirmed the predictive accuracy of these biomarkers both in the training set and test set. Immune cell infiltration analysis suggested that patients with idiopathic pulmonary fibrosis had a higher level of B cells memory, Plasma cells, T cells CD8, T cells follicular helper, T cells regulatory (Tregs), Macrophages M0, and Mast cells resting compared with the control group. Correlation analysis demonstrated that FHL2 was significantly associated with the infiltrating immune cells. qPCR and western blotting analysis suggested that SLAIN1 might be a signature for the diagnosis of idiopathic pulmonary fibrosis. In this study, we identified four potential biomarkers (FHL2, HPCAL1, RNF182, and SLAIN1) and evaluated the potential pathogenic role of SLAIN1 in idiopathic pulmonary fibrosis. These findings may have great significance in guiding the understanding of disease mechanisms and potential therapeutic targets in idiopathic pulmonary fibrosis.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures
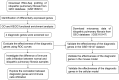

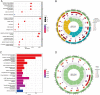





References
-
- Kreuter M, Swigris J, Pittrow D, Geier S, Klotsche J, Prasse A, et al. The clinical course of idiopathic pulmonary fibrosis and its association to quality of life over time: Longitudinal data from the INSIGHTS-IPF registry. Respir. Res. 2019;20(1):1–13. doi: 10.1186/s12931-019-1020-3. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

