Crosstalk between small-cell lung cancer cells and astrocytes mimics brain development to promote brain metastasis
- PMID: 37783795
- PMCID: PMC11230587
- DOI: 10.1038/s41556-023-01241-6
Crosstalk between small-cell lung cancer cells and astrocytes mimics brain development to promote brain metastasis
Abstract
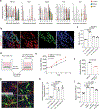
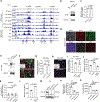
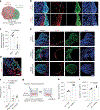
Brain metastases represent an important clinical problem for patients with small-cell lung cancer (SCLC). However, the mechanisms underlying SCLC growth in the brain remain poorly understood. Here, using intracranial injections in mice and assembloids between SCLC aggregates and human cortical organoids in culture, we found that SCLC cells recruit reactive astrocytes to the tumour microenvironment. This crosstalk between SCLC cells and astrocytes drives the induction of gene expression programmes that are similar to those found during early brain development in neurons and astrocytes. Mechanistically, the brain development factor Reelin, secreted by SCLC cells, recruits astrocytes to brain metastases. These astrocytes in turn promote SCLC growth by secreting neuronal pro-survival factors such as SERPINE1. Thus, SCLC brain metastases grow by co-opting mechanisms involved in reciprocal neuron-astrocyte interactions during brain development. Targeting such developmental programmes activated in this cancer ecosystem may help prevent and treat brain metastases.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
J.S. licensed a patent to Forty Seven/Gilead on the use of CD47-blocking strategies in SCLC and has equity in, and is an advisor for, DISCO Pharmaceuticals. M.M.W. has equity in, and is an advisor for, D2G Oncology. M.D. has received recent research support from Novartis, Abbvie, United Therapeutics, Verily and Varian, and has consulted with Beigene, Astra Zeneca and Jazz Pharmaceuticals. M.P. has received honoraria for lectures, consultation or advisory board participation from the following for-profit companies: Bayer, Bristol-Myers Squibb, Novartis, Gerson Lehrman Group (GLG), CMC Contrast, GlaxoSmithKline, Mundipharma, Roche, BMJ Journals, MedMedia, Astra Zeneca, AbbVie, Lilly, Medahead, Daiichi Sankyo, Sanofi, Merck Sharp & Dome, Tocagen, Adastra, Gan & Lee Pharmaceuticals and Servier. The other authors declare no competing interests.
Figures















References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

