Multilevel visual motion opponency in Drosophila
- PMID: 37783895
- PMCID: PMC10620086
- DOI: 10.1038/s41593-023-01443-z
Multilevel visual motion opponency in Drosophila
Abstract
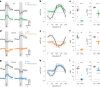
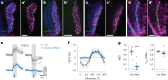
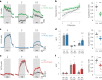
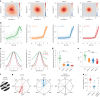
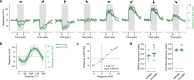
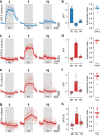
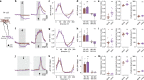
Inhibitory interactions between opponent neuronal pathways constitute a common circuit motif across brain areas and species. However, in most cases, synaptic wiring and biophysical, cellular and network mechanisms generating opponency are unknown. Here, we combine optogenetics, voltage and calcium imaging, connectomics, electrophysiology and modeling to reveal multilevel opponent inhibition in the fly visual system. We uncover a circuit architecture in which a single cell type implements direction-selective, motion-opponent inhibition at all three network levels. This inhibition, mediated by GluClα receptors, is balanced with excitation in strength, despite tenfold fewer synapses. The different opponent network levels constitute a nested, hierarchical structure operating at increasing spatiotemporal scales. Electrophysiology and modeling suggest that distributing this computation over consecutive network levels counteracts a reduction in gain, which would result from integrating large opposing conductances at a single instance. We propose that this neural architecture provides resilience to noise while enabling high selectivity for relevant sensory information.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




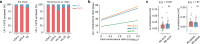


References
-
- Shepherd, G. M. in The Synaptic Organization of the Brain (ed. Shepherd, G. M.) 1–38 (Oxford Univ. Press, 2004).
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

