Epigenomic response to albuterol treatment in asthma-relevant airway epithelial cells
- PMID: 37784136
- PMCID: PMC10546710
- DOI: 10.1186/s13148-023-01571-0
Epigenomic response to albuterol treatment in asthma-relevant airway epithelial cells
Abstract
Background: Albuterol is the first-line asthma medication used in diverse populations. Although DNA methylation (DNAm) is an epigenetic mechanism involved in asthma and bronchodilator drug response (BDR), no study has assessed whether albuterol could induce changes in the airway epithelial methylome. We aimed to characterize albuterol-induced DNAm changes in airway epithelial cells, and assess potential functional consequences and the influence of genetic variation and asthma-related clinical variables.
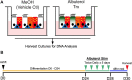
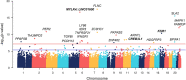
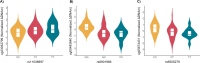
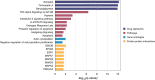
Results: We followed a discovery and validation study design to characterize albuterol-induced DNAm changes in paired airway epithelial cultures stimulated in vitro with albuterol. In the discovery phase, an epigenome-wide association study using paired nasal epithelial cultures from Puerto Rican children (n = 97) identified 22 CpGs genome-wide associated with repeated-use albuterol treatment (p < 9 × 10-8). Albuterol predominantly induced a hypomethylation effect on CpGs captured by the EPIC array across the genome (probability of hypomethylation: 76%, p value = 3.3 × 10-5). DNAm changes on the CpGs cg23032799 (CREB3L1), cg00483640 (MYLK4-LINC01600), and cg05673431 (KSR1) were validated in nasal epithelia from 10 independent donors (false discovery rate [FDR] < 0.05). The effect on the CpG cg23032799 (CREB3L1) was cross-tissue validated in bronchial epithelial cells at nominal level (p = 0.030). DNAm changes in these three CpGs were shown to be influenced by three independent genetic variants (FDR < 0.05). In silico analyses showed these polymorphisms regulated gene expression of nearby genes in lungs and/or fibroblasts including KSR1 and LINC01600 (6.30 × 10-14 ≤ p ≤ 6.60 × 10-5). Additionally, hypomethylation at the CpGs cg10290200 (FLNC) and cg05673431 (KSR1) was associated with increased gene expression of the genes where they are located (FDR < 0.05). Furthermore, while the epigenetic effect of albuterol was independent of the asthma status, severity, and use of medication, BDR was nominally associated with the effect on the CpG cg23032799 (CREB3L1) (p = 0.004). Gene-set enrichment analyses revealed that epigenomic modifications of albuterol could participate in asthma-relevant processes (e.g., IL-2, TNF-α, and NF-κB signaling pathways). Finally, nine differentially methylated regions were associated with albuterol treatment, including CREB3L1, MYLK4, and KSR1 (adjusted p value < 0.05).
Conclusions: This study revealed evidence of epigenetic modifications induced by albuterol in the mucociliary airway epithelium. The epigenomic response induced by albuterol might have potential clinical implications by affecting biological pathways relevant to asthma.
Keywords: Airway cells; Albuterol; CREB3L1; DNA methylation; EWAS; Epigenetics; KSR1; MYLK4; Puerto Ricans; β2-agonist.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
Authors declare they have no competing interests or other interests that might be perceived to influence the interpretation of the article. No supporting institution may gain or lose financially through this publication. The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Figures
References
-
- Global Asthma Network. The Global Asthma Report 2018 [Internet]. Auckland, New Zealand; 2018.
-
- Naqvi M, Thyne S, Choudhry S, Tsai HJ, Navarro D, Castro RA, et al. Ethnic-specific differences in bronchodilator responsiveness among African Americans, Puerto Ricans, and Mexicans with asthma. J Asthma. 2007;44(8):639–648. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 ES015794/ES/NIEHS NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- R01ES015794, R21ES24844/ES/NIEHS NIH HHS/United States
- UM1 HG008901/HG/NHGRI NIH HHS/United States
- R01MD010443, R56MD013312/MD/NIMHD NIH HHS/United States
- R01 HL135156/HL/NHLBI NIH HHS/United States
- R01 HL128439/HL/NHLBI NIH HHS/United States
- R01 HL117004/HL/NHLBI NIH HHS/United States
- R21 ES024844/ES/NIEHS NIH HHS/United States
- R01 HL117626/HL/NHLBI NIH HHS/United States
- U24 HG008956/HG/NHGRI NIH HHS/United States
- R56 MD013312/MD/NIMHD NIH HHS/United States
- R01 MD010443/MD/NIMHD NIH HHS/United States
- HHSN268201600032C/ES/NIEHS NIH HHS/United States
- R01 HL155024/HL/NHLBI NIH HHS/United States
- R01HL155024-01, HHSN268201600032I, 3R01HL-117626-02S1, HHSN268201800002I, 3R01HL117004-02S3, 3R01HL-120393-02S1, R01HL117004, R01HL128439, R01HL135156, X01HL134589/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

