QuTIE: quantum optimization for target identification by enzymes
- PMID: 37786534
- PMCID: PMC10541652
- DOI: 10.1093/bioadv/vbad112
QuTIE: quantum optimization for target identification by enzymes
Abstract
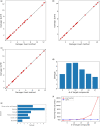
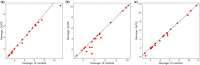
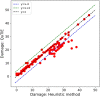
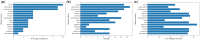
Summary: Target identification by enzymes (TIE) problem aims to identify the set of enzymes in a given metabolic network, such that their inhibition eliminates a given set of target compounds associated with a disease while incurring minimum damage to the rest of the compounds. This is a NP-hard problem, and thus optimal solutions using classical computers fail to scale to large metabolic networks. In this article, we develop the first quantum optimization solution, called QuTIE (quantum optimization for target identification by enzymes), to this NP-hard problem. We do that by developing an equivalent formulation of the TIE problem in quadratic unconstrained binary optimization form. We then map it to a logical graph, and embed the logical graph on a quantum hardware graph. Our experimental results on 27 metabolic networks from Escherichia coli, Homo sapiens, and Mus musculus show that QuTIE yields solutions that are optimal or almost optimal. Our experiments also demonstrate that QuTIE can successfully identify enzyme targets already verified in wet-lab experiments for 14 major disease classes.
Availability and implementation: Code and sample data are available at: https://github.com/ngominhhoang/Quantum-Target-Identification-by-Enzymes.
Published by Oxford University Press 2023.
Conflict of interest statement
None declared.
Figures

References
-
- Barse AV, Chakrabarti T, Ghosh TK. et al. Endocrine disruption and metabolic changes following exposure of Cyprinus carpio to diethyl phthalate. Pestic Biochem Physiol 2007;88:36–42.
-
- Choi V. Minor-embedding in adiabatic quantum computation: I. The parameter setting problem. Quantum Inf Process 2008;7:193–209.
-
- Cohen J, Powderly W, Opal S.. Infectious Diseases. 3rd edn. Amsterdam, Netherlands: Elsevier Inc., 2010.
LinkOut - more resources
Full Text Sources
Miscellaneous

