Genome-Wide Association Analysis of Protein-Coding Variants in IgA Nephropathy
- PMID: 37787447
- PMCID: PMC10631603
- DOI: 10.1681/ASN.0000000000000222
Genome-Wide Association Analysis of Protein-Coding Variants in IgA Nephropathy
Abstract
Significance statement: Genome-wide association studies have identified nearly 20 IgA nephropathy susceptibility loci. However, most nonsynonymous coding variants, particularly ones that occur rarely or at a low frequency, have not been well investigated. The authors performed a chip-based association study of IgA nephropathy in 8529 patients with the disorder and 23,224 controls. They identified a rare variant in the gene encoding vascular endothelial growth factor A (VEGFA) that was significantly associated with a two-fold increased risk of IgA nephropathy, which was further confirmed by sequencing analysis. They also identified a novel common variant in PKD1L3 that was significantly associated with lower haptoglobin protein levels. This study, which was well-powered to detect low-frequency variants with moderate to large effect sizes, helps expand our understanding of the genetic basis of IgA nephropathy susceptibility.
Background: Genome-wide association studies have identified nearly 20 susceptibility loci for IgA nephropathy. However, most nonsynonymous coding variants, particularly those occurring rarely or at a low frequency, have not been well investigated.
Methods: We performed a three-stage exome chip-based association study of coding variants in 8529 patients with IgA nephropathy and 23,224 controls, all of Han Chinese ancestry. Sequencing analysis was conducted to investigate rare coding variants that were not covered by the exome chip. We used molecular dynamic simulation to characterize the effects of mutations of VEGFA on the protein's structure and function. We also explored the relationship between the identified variants and the risk of disease progression.
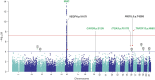
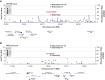
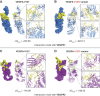
Results: We discovered a novel rare nonsynonymous risk variant in VEGFA (odds ratio, 1.97; 95% confidence interval [95% CI], 1.61 to 2.41; P = 3.61×10 -11 ). Further sequencing of VEGFA revealed twice as many carriers of other rare variants in 2148 cases compared with 2732 controls. We also identified a common nonsynonymous risk variant in PKD1L3 (odds ratio, 1.16; 95% CI, 1.11 to 1.21; P = 1.43×10 -11 ), which was associated with lower haptoglobin protein levels. The rare VEGFA mutation could cause a conformational change and increase the binding affinity of VEGFA to its receptors. Furthermore, this variant was associated with the increased risk of kidney disease progression in IgA nephropathy (hazard ratio, 2.99; 95% CI, 1.09 to 8.21; P = 0.03).
Conclusions: Our study identified two novel risk variants for IgA nephropathy in VEGFA and PKD1L3 and helps expand our understanding of the genetic basis of IgA nephropathy susceptibility.
Copyright © 2023 by the American Society of Nephrology.
Conflict of interest statement
J.-X. Bei reports Ownership Interest: Junshi Ltd.; and Patents or Royalties: Sun Yat-sen University Cancer Center. G.-R. Jiang reports Research Funding: Fibrogen. J. Lee reports Consultancy: Boehringer Ingelheim; and Advisory or Leadership Role: ThoughtFull World Pte. Ltd. E.S. Tai reports Consultancy: Farma Mondo S.A., Novartis Singapore Pte. Ltd., and Kowa Pharmaceutical Asia Pte. Ltd.; Ownership Interest: Abbott, Abbvie, and Novavax; Honoraria: As listed under consultancy agreements; and Speakers Bureau: as listed under consultancy agreements. E.-K. Tan reports Research Funding: YiQi Company; and Honoraria: Academic activities (Esai) and Editorial duties (Elsevier). H. Zhang reports Consultancy: Calliditas, Chinook, Novartis, OMEROS, and Ostuka; and Advisory or Leadership Role: Ostuka. All remaining authors have nothing to disclose.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

