The semaphorin 3A/neuropilin-1 pathway promotes clonogenic growth of glioblastoma via activation of TGF-β signaling
- PMID: 37788099
- PMCID: PMC10721275
- DOI: 10.1172/jci.insight.167049
The semaphorin 3A/neuropilin-1 pathway promotes clonogenic growth of glioblastoma via activation of TGF-β signaling
Abstract
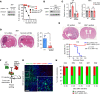
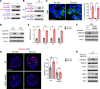
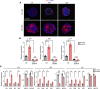
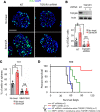
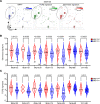
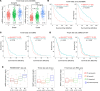
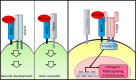
Glioblastoma (GBM) is the most lethal brain cancer with a dismal prognosis. Stem-like GBM cells (GSCs) are a major driver of GBM propagation and recurrence; thus, understanding the molecular mechanisms that promote GSCs may lead to effective therapeutic approaches. Through in vitro clonogenic growth-based assays, we determined mitogenic activities of the ligand molecules that are implicated in neural development. We have identified that semaphorin 3A (Sema3A), originally known as an axon guidance molecule in the CNS, promotes clonogenic growth of GBM cells but not normal neural progenitor cells (NPCs). Mechanistically, Sema3A binds to its receptor neuropilin-1 (NRP1) and facilitates an interaction between NRP1 and TGF-β receptor 1 (TGF-βR1), which in turn leads to activation of canonical TGF-β signaling in both GSCs and NPCs. TGF-β signaling enhances self-renewal and survival of GBM tumors through induction of key stem cell factors, but it evokes cytostatic responses in NPCs. Blockage of the Sema3A/NRP1 axis via shRNA-mediated knockdown of Sema3A or NRP1 impeded clonogenic growth and TGF-β pathway activity in GSCs and inhibited tumor growth in vivo. Taken together, these findings suggest that the Sema3A/NRP1/TGF-βR1 signaling axis is a critical regulator of GSC propagation and a potential therapeutic target for GBM.
Keywords: Brain cancer; Development; Growth factors; Oncogenes; Oncology.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

