AMPK inhibitor, compound C, inhibits coronavirus replication in vitro
- PMID: 37788269
- PMCID: PMC10547180
- DOI: 10.1371/journal.pone.0292309
AMPK inhibitor, compound C, inhibits coronavirus replication in vitro
Abstract
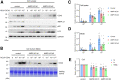
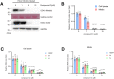
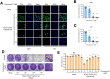
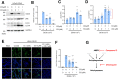
The coronavirus disease (COVID-19) pandemic has resulted in more than six million deaths by October 2022. Vaccines and antivirals for severe acute respiratory syndrome coronavirus 2 are now available; however, more effective antiviral drugs are required for effective treatment. Here, we report that a potent AMP-activated protein kinase (AMPK) inhibitor, compound C/dorsomorphin, inhibits the replication of the human coronavirus OC43 strain (HCoV-OC43). We examined HCoV-OC43 replication in control and AMPK-knockout (KO) cells and found that the virus replication decreased in AMPK-KO cells. Next, we examined the effect of the AMPK inhibitor, compound C on coronavirus replication. Compound C treatment efficiently inhibited the replication and decreased the coronavirus-induced cytotoxicity, further inhibiting autophagy. In addition, treatment with compound C in combination with chloroquine synergistically inhibited coronavirus replication. These results suggest that compound C can be considered as a potential drug candidate for COVID-19.
Copyright: © 2023 Jang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
NO authors have competing interests.
Figures
Similar articles
-
Antiviral activity of chloroquine against human coronavirus OC43 infection in newborn mice.Antimicrob Agents Chemother. 2009 Aug;53(8):3416-21. doi: 10.1128/AAC.01509-08. Epub 2009 Jun 8. Antimicrob Agents Chemother. 2009. PMID: 19506054 Free PMC article.
-
EGCG, a green tea polyphenol, inhibits human coronavirus replication in vitro.Biochem Biophys Res Commun. 2021 Apr 2;547:23-28. doi: 10.1016/j.bbrc.2021.02.016. Epub 2021 Feb 10. Biochem Biophys Res Commun. 2021. PMID: 33588235 Free PMC article.
-
Safe and Sensitive Antiviral Screening Platform Based on Recombinant Human Coronavirus OC43 Expressing the Luciferase Reporter Gene.Antimicrob Agents Chemother. 2016 Aug 22;60(9):5492-503. doi: 10.1128/AAC.00814-16. Print 2016 Sep. Antimicrob Agents Chemother. 2016. PMID: 27381385 Free PMC article.
-
Antiviral strategies against human coronaviruses.Infect Disord Drug Targets. 2007 Mar;7(1):59-66. doi: 10.2174/187152607780090757. Infect Disord Drug Targets. 2007. PMID: 17346212 Review.
-
Human Coronavirus OC43 as a Low-Risk Model to Study COVID-19.Viruses. 2023 Feb 20;15(2):578. doi: 10.3390/v15020578. Viruses. 2023. PMID: 36851792 Free PMC article. Review.
Cited by
-
Amentoflavone from Selaginella tamariscina inhibits SARS-CoV-2 RNA-dependent RNA polymerase.Heliyon. 2024 Aug 20;10(16):e36568. doi: 10.1016/j.heliyon.2024.e36568. eCollection 2024 Aug 30. Heliyon. 2024. PMID: 39258186 Free PMC article.
-
Astaxanthin Alleviates Oxidative Stress in Mouse Preantral Follicles and Enhances Follicular Development Through the AMPK Signaling Pathway.Int J Mol Sci. 2025 Mar 2;26(5):2241. doi: 10.3390/ijms26052241. Int J Mol Sci. 2025. PMID: 40076863 Free PMC article.
-
Arctigenin from Forsythia viridissima Fruit Inhibits the Replication of Human Coronavirus.Int J Mol Sci. 2024 Jul 4;25(13):7363. doi: 10.3390/ijms25137363. Int J Mol Sci. 2024. PMID: 39000469 Free PMC article.
-
AMPK Knockout Impairs the Formation of Three-Dimensional Spheroids.Life (Basel). 2025 Mar 22;15(4):525. doi: 10.3390/life15040525. Life (Basel). 2025. PMID: 40283080 Free PMC article.
-
Chrysanthemum zawadskii ethanol extract inhibits the replication of alpha-coronavirus and beta-coronavirus.PLoS One. 2025 Jun 18;20(6):e0326225. doi: 10.1371/journal.pone.0326225. eCollection 2025. PLoS One. 2025. PMID: 40531892 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

