A dominant negative 14-3-3 mutant in Schizosaccharomyces pombe distinguishes the binding proteins involved in sexual differentiation and check point
- PMID: 37788281
- PMCID: PMC10547172
- DOI: 10.1371/journal.pone.0291524
A dominant negative 14-3-3 mutant in Schizosaccharomyces pombe distinguishes the binding proteins involved in sexual differentiation and check point
Abstract
The homothallic fission yeast Schizosaccharomyces pombe undergoes sexual differentiation when starved, but sam (skips the requirement of starvation for mating) mutants such as those carrying mutations in adenylate cyclase (cyr1) or protein kinase A (pka1) mate without starvation. Here, we identified sam3, a dominant negative allele of rad24, encoding one of two 14-3-3 proteins. Genetic mapping and whole-genome sequencing showed that the sam3 mutation comprises a change in nucleotide at position 959 from guanine to adenine, which switches the amino acid at position 185 from glutamic acid to lysine (E185K). We generated the rad24-E185K integrated mutant and its phenotype was similar to that of the sam3 mutant, including calcium sensitivity and UV non-sensitivity, but the phenotype is different from that of the Δrad24 strain. While the UV-sensitive phenotype was observed in the Δrad24 mutant, it was not observed in the sam3 and rad24-E185K mutants. The expression of the rad24-E185K gene in wild type cells induced spore formation in the nutrient rich medium, confirming rad24-E185K is dominant. This dominant effect of rad24-E185K was also observed in Δras1 and Δbyr2 diploid mutants, indicating that rad24-E185K generate stronger phenotype than rad24 null mutants. Ste11, the key transcription factor for sexual differentiation was expressed in sam3 mutants without starvation and it predominantly localized to the nucleus. The Rad24-E185K mutant protein retained its interaction with Check point kinase1 (Chk1), whereas it reduced interaction with Ste11, an RNA binding protein Mei2, and a MAPKKK Byr2, freeing these proteins from negative regulation by Rad24, that account for the sam phenotype and UV non-sensitive phenotype. Glucose depletion in rad24-E185K or Δpka1 Δrad24 double mutation induced haploid meiosis, leading to the formation of spores in haploid. The position of glutamic acid 185 is conserved in all major 14-3-3s; hence, our finding of a dominant negative allele of 14-3-3 is useful for understanding 14-3-3s in other organisms.
Copyright: © 2023 Ohshima et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



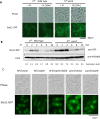

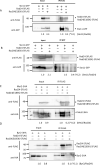
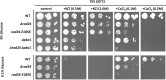
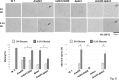

Similar articles
-
The 14-3-3 proteins Rad24 and Rad25 negatively regulate Byr2 by affecting its localization in Schizosaccharomyces pombe.Mol Cell Biol. 2002 Oct;22(20):7105-19. doi: 10.1128/MCB.22.20.7105-7119.2002. Mol Cell Biol. 2002. PMID: 12242289 Free PMC article.
-
Identification of sam4 as a rad24 allele in Schizosaccharomyces pombe.Biosci Biotechnol Biochem. 2009 Jul;73(7):1591-8. doi: 10.1271/bbb.90103. Epub 2009 Jul 7. Biosci Biotechnol Biochem. 2009. PMID: 19584544
-
Rad24 is essential for proliferation of diploid cells in fission yeast.FEBS Lett. 2000 Apr 28;472(2-3):254-8. doi: 10.1016/s0014-5793(00)01395-8. FEBS Lett. 2000. PMID: 10788621
-
Regulation of meiosis in fission yeast.Cell Struct Funct. 1996 Oct;21(5):431-6. doi: 10.1247/csf.21.431. Cell Struct Funct. 1996. PMID: 9118252 Review.
-
Analysis of Schizosaccharomyces pombe Meiosis.Cold Spring Harb Protoc. 2017 Sep 1;2017(9):pdb.top079855. doi: 10.1101/pdb.top079855. Cold Spring Harb Protoc. 2017. PMID: 28733417 Review.
Cited by
-
Mutational analyses of the interacting domains of Schizosaccharomyces pombe Byr2 with 14-3-3s.Curr Genet. 2024 Jun 24;70(1):8. doi: 10.1007/s00294-024-01293-7. Curr Genet. 2024. PMID: 38913087 Free PMC article.
References
-
- Goldar MM, Nishie T, Ishikura Y, Fukuda T, Takegawa K, Kawamukai M. Functional conservation between fission yeast moc1/sds23 and its two orthologs, budding yeast SDS23 and SDS24, and phenotypic differences in their disruptants. Bioscience Biotechnology and Biochemistry. 2005;69(7):1422–6. doi: 10.1271/bbb.69.1422 WOS:000231055700031. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

