The duplication of genomes and genetic networks and its potential for evolutionary adaptation and survival during environmental turmoil
- PMID: 37788315
- PMCID: PMC10576144
- DOI: 10.1073/pnas.2307289120
The duplication of genomes and genetic networks and its potential for evolutionary adaptation and survival during environmental turmoil
Abstract
The importance of whole-genome duplication (WGD) for evolution is controversial. Whereas some view WGD mainly as detrimental and an evolutionary dead end, there is growing evidence that polyploidization can help overcome environmental change, stressful conditions, or periods of extinction. However, despite much research, the mechanistic underpinnings of why and how polyploids might be able to outcompete or outlive nonpolyploids at times of environmental upheaval remain elusive, especially for autopolyploids, in which heterosis effects are limited. On the longer term, WGD might increase both mutational and environmental robustness due to redundancy and increased genetic variation, but on the short-or even immediate-term, selective advantages of WGDs are harder to explain. Here, by duplicating artificially generated Gene Regulatory Networks (GRNs), we show that duplicated GRNs-and thus duplicated genomes-show higher signal output variation than nonduplicated GRNs. This increased variation leads to niche expansion and can provide polyploid populations with substantial advantages to survive environmental turmoil. In contrast, under stable environments, GRNs might be maladaptive to changes, a phenomenon that is exacerbated in duplicated GRNs. We believe that these results provide insights into how genome duplication and (auto)polyploidy might help organisms to adapt quickly to novel conditions and to survive ecological uproar or even cataclysmic events.
Keywords: cataclysmic events; environmental turmoil; gene regulatory networks; polyploidy; whole-genome duplication.
Conflict of interest statement
The authors declare no competing interest.
Figures
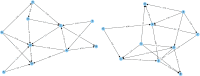
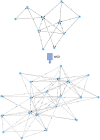

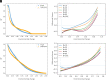
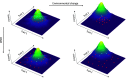
Similar articles
-
Using digital organisms to study the evolutionary consequences of whole genome duplication and polyploidy.PLoS One. 2019 Jul 31;14(7):e0220257. doi: 10.1371/journal.pone.0220257. eCollection 2019. PLoS One. 2019. PMID: 31365541 Free PMC article.
-
Whole-genome Duplications and the Long-term Evolution of Gene Regulatory Networks in Angiosperms.Mol Biol Evol. 2023 Jul 5;40(7):msad141. doi: 10.1093/molbev/msad141. Mol Biol Evol. 2023. PMID: 37405949 Free PMC article.
-
Polyploidy: an evolutionary and ecological force in stressful times.Plant Cell. 2021 Mar 22;33(1):11-26. doi: 10.1093/plcell/koaa015. Plant Cell. 2021. PMID: 33751096 Free PMC article. Review.
-
Polyploidy and genome evolution in plants.Curr Opin Genet Dev. 2015 Dec;35:119-25. doi: 10.1016/j.gde.2015.11.003. Epub 2015 Dec 2. Curr Opin Genet Dev. 2015. PMID: 26656231 Review.
-
wgd v2: a suite of tools to uncover and date ancient polyploidy and whole-genome duplication.Bioinformatics. 2024 May 2;40(5):btae272. doi: 10.1093/bioinformatics/btae272. Bioinformatics. 2024. PMID: 38632086 Free PMC article.
Cited by
-
Parallel Selection in Domesticated Atlantic Salmon from Divergent Founders Including on Whole-Genome Duplication-derived Homeologous Regions.Genome Biol Evol. 2025 Apr 3;17(4):evaf063. doi: 10.1093/gbe/evaf063. Genome Biol Evol. 2025. PMID: 40247730 Free PMC article.
-
Expression pattern changes of three homeologs in chemokine activity enhance antiviral response to herpesvirus infection in a newly synthesized alloheptaploid.BMC Genomics. 2025 Jul 14;26(1):662. doi: 10.1186/s12864-025-11838-w. BMC Genomics. 2025. PMID: 40653463 Free PMC article.
-
The immediate metabolomic effects of whole-genome duplication in the greater duckweed, Spirodela polyrhiza.Am J Bot. 2024 Aug;111(8):e16383. doi: 10.1002/ajb2.16383. Epub 2024 Aug 1. Am J Bot. 2024. PMID: 39087852 Free PMC article.
-
Neopolyploidy increases stress tolerance and reduces fitness plasticity across multiple urban pollutants: support for the "general-purpose" genotype hypothesis.Evol Lett. 2024 Jan 10;8(3):416-426. doi: 10.1093/evlett/qrad072. eCollection 2024 Jun. Evol Lett. 2024. PMID: 38818423 Free PMC article.
-
Reticulate allopolyploidy and subsequent dysploidy drive evolution and diversification in the cotton family.Nat Commun. 2025 Aug 12;16(1):7480. doi: 10.1038/s41467-025-62644-7. Nat Commun. 2025. PMID: 40796551 Free PMC article.
References
-
- Comai L., The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846 (2005). - PubMed
-
- Levin D. A., Minority cytotype exclusion in local plant populations. Taxon 24, 35–43 (1975).
-
- Wendel J. F., The wondrous cycles of polyploidy in plants. Am. J. Bot. 102, 1753–1756 (2015). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

