Structural insights into the conformational changes of BTR1/SLC4A11 in complex with PIP2
- PMID: 37788993
- PMCID: PMC10547724
- DOI: 10.1038/s41467-023-41924-0
Structural insights into the conformational changes of BTR1/SLC4A11 in complex with PIP2
Abstract
BTR1 (SLC4A11) is a NH3 stimulated H+ (OH-) transporter belonging to the SLC4 family. Dysfunction of BTR1 leads to diseases such as congenital hereditary endothelial dystrophy (CHED) and Fuchs endothelial corneal dystrophy (FECD). However, the mechanistic basis of BTR1 activation by alkaline pH, transport activity regulation and pathogenic mutations remains elusive. Here, we present cryo-EM structures of human BTR1 in the outward-facing state in complex with its activating ligands PIP2 and the inward-facing state with the pathogenic R125H mutation. We reveal that PIP2 binds at the interface between the transmembrane domain and the N-terminal cytosolic domain of BTR1. Disruption of either the PIP2 binding site or protonation of PIP2 phosphate groups by acidic pH can transform BTR1 into an inward-facing conformation. Our results provide insights into the mechanisms of how the transport activity and conformation changes of BTR1 are regulated by PIP2 binding and interaction of TMD and NTD.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures





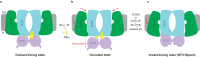
Similar articles
-
Coexistence of Congenital Hereditary Endothelial Dystrophy and Fuchs Endothelial Corneal Dystrophy Associated With SLC4A11 Mutations in Affected Families.Cornea. 2020 Mar;39(3):354-357. doi: 10.1097/ICO.0000000000002183. Cornea. 2020. PMID: 31714402
-
Fuchs Endothelial Corneal Dystrophy in a Heterozygous Carrier of Congenital Hereditary Endothelial Dystrophy Type 2 with a Novel Mutation in SLC4A11.Ophthalmic Genet. 2015;36(3):284-6. doi: 10.3109/13816810.2014.881510. Ophthalmic Genet. 2015. PMID: 24502824
-
Observation of nine previously reported and 10 non-reported SLC4A11 mutations among 20 Iranian CHED probands and identification of an MPDZ mutation as possible cause of CHED and FECD in one family.Br J Ophthalmol. 2020 Nov;104(11):1621-1628. doi: 10.1136/bjophthalmol-2019-314377. Epub 2019 Aug 16. Br J Ophthalmol. 2020. PMID: 31420327
-
Update on the genetics of corneal endothelial dystrophies.Indian J Ophthalmol. 2022 Jul;70(7):2239-2248. doi: 10.4103/ijo.IJO_992_22. Indian J Ophthalmol. 2022. PMID: 35791103 Free PMC article. Review.
-
SLC4A11 and the Pathophysiology of Congenital Hereditary Endothelial Dystrophy.Biomed Res Int. 2015;2015:475392. doi: 10.1155/2015/475392. Epub 2015 Sep 16. Biomed Res Int. 2015. PMID: 26451371 Free PMC article. Review.
Cited by
-
Abnormal Cellular Populations Shape Thymic Epithelial Tumor Heterogeneity and Anti-Tumor by Blocking Metabolic Interactions in Organoids.Adv Sci (Weinh). 2024 Nov;11(42):e2406653. doi: 10.1002/advs.202406653. Epub 2024 Sep 11. Adv Sci (Weinh). 2024. PMID: 39258580 Free PMC article.
-
On the substrate turnover rate of NBCe1 and AE1 SLC4 transporters: structure-function considerations.Front Physiol. 2025 Jan 13;15:1474628. doi: 10.3389/fphys.2024.1474628. eCollection 2024. Front Physiol. 2025. PMID: 39872416 Free PMC article.
-
A role for pH dynamics regulating transcription factor DNA-binding selectivity.Nucleic Acids Res. 2025 May 22;53(10):gkaf474. doi: 10.1093/nar/gkaf474. Nucleic Acids Res. 2025. PMID: 40464693 Free PMC article.
-
Structural and functional insights into the lipid regulation of human anion exchanger 2.Nat Commun. 2024 Jan 26;15(1):759. doi: 10.1038/s41467-024-44966-0. Nat Commun. 2024. PMID: 38272905 Free PMC article.
-
Memorization Bias Impacts Modeling of Alternative Conformational States of Symmetric Solute Carrier Membrane Proteins with Methods from Deep Learning.bioRxiv [Preprint]. 2025 Apr 26:2024.07.15.603529. doi: 10.1101/2024.07.15.603529. bioRxiv. 2025. PMID: 39071413 Free PMC article. Preprint.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

