Effects of tea, catechins and catechin derivatives on Omicron subvariants of SARS-CoV-2
- PMID: 37789046
- PMCID: PMC10547759
- DOI: 10.1038/s41598-023-43563-3
Effects of tea, catechins and catechin derivatives on Omicron subvariants of SARS-CoV-2
Abstract
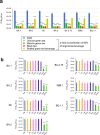
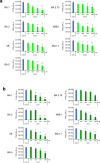
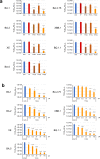
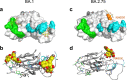
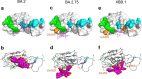
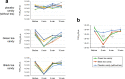
The Omicron subvariants of SARS-CoV-2 have multiple mutations in the S-proteins and show high transmissibility. We previously reported that tea catechin (-)-epigallocatechin gallate (EGCG) and its derivatives including theaflavin-3,3'-di-O-digallate (TFDG) strongly inactivated the conventional SARS-CoV-2 by binding to the receptor binding domain (RBD) of the S-protein. Here we show that Omicron subvariants were effectively inactivated by green tea, Matcha, and black tea. EGCG and TFDG strongly suppressed infectivity of BA.1 and XE subvariants, while effect on BA.2.75 was weaker. Neutralization assay showed that EGCG and TFDG inhibited interaction between BA.1 RBD and ACE2. In silico analyses suggested that N460K, G446S and F490S mutations in RBDs crucially influenced the binding of EGCG/TFDG to the RBDs. Healthy volunteers consumed a candy containing green tea or black tea, and saliva collected from them immediately after the candy consumption significantly decreased BA.1 virus infectivity in vitro. These results indicate specific amino acid substitutions in RBDs that crucially influence the binding of EGCG/TFDG to the RBDs and different susceptibility of each Omicron subvariant to EGCG/TFDG. The study may suggest molecular basis for potential usefulness of these compounds in suppression of mutant viruses that could emerge in the future and cause next pandemic.
© 2023. Springer Nature Limited.
Conflict of interest statement
O.M. has received funding support from Japan Science and Technology Agency and ITO EN, ltd, Tokyo, Japan. Other authors declare no COI. The company also provided information of tea, catechins and catechin derivatives, and discussion with authors. The funders had no role in the design of the study, in the interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

