An eXplainable Artificial Intelligence analysis of Raman spectra for thyroid cancer diagnosis
- PMID: 37789191
- PMCID: PMC10547772
- DOI: 10.1038/s41598-023-43856-7
An eXplainable Artificial Intelligence analysis of Raman spectra for thyroid cancer diagnosis
Abstract
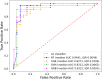
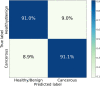
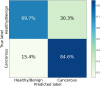
Raman spectroscopy shows great potential as a diagnostic tool for thyroid cancer due to its ability to detect biochemical changes during cancer development. This technique is particularly valuable because it is non-invasive and label/dye-free. Compared to molecular tests, Raman spectroscopy analyses can more effectively discriminate malignant features, thus reducing unnecessary surgeries. However, one major hurdle to using Raman spectroscopy as a diagnostic tool is the identification of significant patterns and peaks. In this study, we propose a Machine Learning procedure to discriminate healthy/benign versus malignant nodules that produces interpretable results. We collect Raman spectra obtained from histological samples, select a set of peaks with a data-driven and label independent approach and train the algorithms with the relative prominence of the peaks in the selected set. The performance of the considered models, quantified by area under the Receiver Operating Characteristic curve, exceeds 0.9. To enhance the interpretability of the results, we employ eXplainable Artificial Intelligence and compute the contribution of each feature to the prediction of each sample.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- NIH National Cancer Institute. Thyroid Cancer—Cancer Stat Facts. https://seer.cancer.gov/statfacts/html/thyro.html (2023). Accessed 22 June 2023.
-
- Vaccarella S, et al. Worldwide thyroid-cancer epidemic? The increasing impact of overdiagnosis. N. Engl. J. Med. 2016;375:614–617. - PubMed
-
- Patel KN, et al. The American Association of Endocrine Surgeons guidelines for the definitive surgical management of thyroid disease in adults. Ann. Surg. 2020;271:e21–e93. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

