LINC00922 decoys SIRT3 to facilitate the metastasis of colorectal cancer through up-regulation the H3K27 crotonylation of ETS1 promoter
- PMID: 37789393
- PMCID: PMC10548613
- DOI: 10.1186/s12943-023-01859-y
LINC00922 decoys SIRT3 to facilitate the metastasis of colorectal cancer through up-regulation the H3K27 crotonylation of ETS1 promoter
Abstract
Background: Lysine crotonylation (Kcr) is up-regulation in colorectal cancer (CRC) tissues, while its specific contribution remains uncertain. This study aimed to elucidate the role and mechanism of crotonylation on Lys27 of histone H3 (H3K27cr) in facilitating CRC metastasis.
Methods: Immunohistochemistry was employed to investigate the correlation between H3K27cr and CRC metastasis. Both in vitro and in vivo assays employing loss function or gain function approaches were conducted to elucidate the role of LINC00922 in promoting CRC metastasis. ScRNA-seq analysis and immunoprecipitation analyses were employed to explore the underlying mechanism by which LINC00922 facilitates CRC metastasis through H3K27cr.
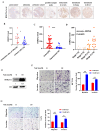
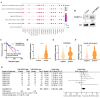
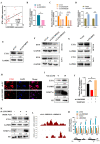
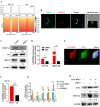
Results: Clinically, H3K27cr was upregulated in metastatic CRC tissues and positively correlated with advanced clinical stages. Functionally, knockdown of LINC00922 inhibited migration of CRC cells both in vitro and in vivo. Furthermore, the supplementation of NaCr restored the migration and invasion levels of LINC00922 stable knockdown cells by restoring the H3K27cr level. Mechanistically, LINC00922 promoted invasion and migration through H3K27cr mediated cell adhesion molecules (CAMs) in epithelial cells. Notably, LINC00922 interacted with the protein sirtuin 3 (SIRT3) and obstructed its binding to the promoter region of ETS1, leading to an elevation in the level of H3K27cr in this promoter region and the subsequent activation of ETS1 transcription.
Conclusions: Our findings uncovered a novel regulatory function of H3K27cr, regulated by LINC00922, in facilitating CRC metastasis. This discovery contributed to a deeper comprehension of the involvement of histone crotonylation in the metastatic process of CRC.
Keywords: Colorectal cancer; H3K27cr; LINC00922; Metastasis; SIRT3.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
The authors declare no potential conflicts of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

