The PyPIF5-PymiR156a-PySPL9-PyMYB114/MYB10 module regulates light-induced anthocyanin biosynthesis in red pear
- PMID: 37789406
- PMCID: PMC10514999
- DOI: 10.1186/s43897-021-00018-5
The PyPIF5-PymiR156a-PySPL9-PyMYB114/MYB10 module regulates light-induced anthocyanin biosynthesis in red pear
Abstract
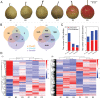
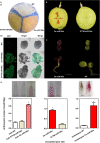
Some cultivars of pear (Pyrus L.) show attractive red fruit skin due to anthocyanin accumulation. This pigmentation can be affected by environmental conditions, especially light. To explore the light-induced regulation network for anthocyanin biosynthesis and fruit coloration in pear, small RNA libraries and mRNA libraries from fruit skins of 'Yunhongyihao' pear were constructed to compare the difference between bagging and debagging treatments. Analysis of RNA-seq of fruit skins with limited light (bagged) and exposed to light (debagged), showed that PyPIF5 was down-regulated after bag removal. PymiR156a was also differentially expressed between bagged and debagged fruit skins. We found that PyPIF5 negatively regulated PymiR156a expression in bagged fruits by directly binding to the G-box motif in its promoter. In addition, PymiR156a overexpression promoted anthocyanin accumulation in both pear skin and apple calli. We confirmed that PymiR156a mediated the cleavage of PySPL9, and that the target PySPL9 protein could form heterodimers with two key anthocyanin regulators (PyMYB114/PyMYB10). We proposed a new module of PyPIF5-PymiR156a-PySPL9-PyMYB114/MYB10. When the bagged fruits were re-exposed to light, PyPIF5 was down-regulated and its inhibitory effect on PymiR156a was weakened, which leads to degradation of the target PySPL, thus eliminating the blocking effect of PySPL on the formation of the regulatory MYB complexes. Ultimately, this promotes anthocyanin biosynthesis in pear skin.
Keywords: Anthocyanin; PyMYB114/MYB10; PyPIF5; PySPL9; PymiR156a; Red-skinned pear.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
