Decoding stoichiometric protein synthesis in E. coli through translation rate parameters
- PMID: 37789867
- PMCID: PMC10542608
- DOI: 10.1016/j.bpr.2023.100131
Decoding stoichiometric protein synthesis in E. coli through translation rate parameters
Abstract
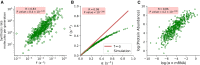
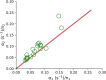
E. coli is one of the most widely used organisms for understanding the principles of cellular and molecular genetics. However, we are yet to understand the origin of several experimental observations related to the regulation of gene expression in E. coli. One of the prominent examples in this context is the proportional synthesis in multiprotein complexes where all of their obligate subunits are produced in proportion to their stoichiometry. In this work, by combining the next-generation sequencing data with the stochastic simulations of protein synthesis, we explain the origin of proportional protein synthesis in multicomponent complexes. We find that the estimated initiation rates for the translation of all subunits in those complexes are proportional to their stoichiometry. This constraint on protein synthesis kinetics enforces proportional protein synthesis without requiring any feedback mechanism. We also find that the translation initiation rates in E. coli are influenced by the coding sequence length and the enrichment of A and C nucleotides near the start codon. Thus, this study rationalizes the role of conserved and nonrandom features of genes in regulating the translation kinetics and unravels a key principle of the regulation of protein synthesis.
© 2023 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Schliwa M., Woehlke G. Molecular motors. Nature. 2003;422:759–765. - PubMed
-
- Martin G.S. Cell signaling and cancer. Cancer Cell. 2003;4:167–174. - PubMed
-
- Møller J.V., Juul B., le Maire M. Structural organization, ion transport, and energy transduction of p-type atpases. Biochim. Biophys. Acta. 1996;1286:1–51. - PubMed
LinkOut - more resources
Full Text Sources

