Novel variants in the CLCN4 gene associated with syndromic X-linked intellectual disability
- PMID: 37789889
- PMCID: PMC10542403
- DOI: 10.3389/fneur.2023.1096969
Novel variants in the CLCN4 gene associated with syndromic X-linked intellectual disability
Abstract
Objective: The dysfunction of the CLCN4 gene can lead to X-linked intellectual disability and Raynaud-Claes syndrome (MRXSRC), characterized by severe cognitive impairment and mental disorders. This study aimed to investigate the genetic defects and clinical features of Chinese children with CLCN4 variants and explore the effect of mutant ClC-4 on the protein expression level and subcellular localization through in vitro experiments.
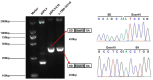
Methods: A total of 401 children with intellectual disabilities were screened for genetic variability using whole-exome sequencing (WES). Clinical data, including age, sex, perinatal conditions, and environmental exposure, were collected. Cognitive, verbal, motor, and social behavioral abilities were evaluated. Candidate variants were verified using Sanger sequencing, and their pathogenicity and conservation were analyzed using in silico prediction tools. Protein expression and localization of mutant ClC-4 were measured using Western blotting (WB) and immunofluorescence microscopy. The impact of a splice site variant was assessed with a minigene assay.
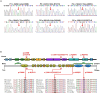
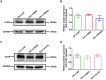
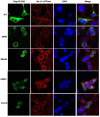
Results: Exome analysis identified five rare CLCN4 variants in six unrelated patients with intellectual disabilities, including two recurrent heterozygous de novo missense variants (p.D89N and p.A555V) in three female patients, and two hemizygous missense variants (p.N141S and p.R694Q) and a splicing variant (c.1390-12T > G) that are maternally inherited in three male patients. The p.N141S variant and the splicing variant c.1390-12(T > G were novel, while p.R694Q was identified in two asymptomatic heterozygous female patients. The six children with CLCN4 variants exhibited a neurodevelopmental spectrum disease characterized by intellectual disability (ID), delayed speech, autism spectrum disorders (ASD), microcephaly, hypertonia, and abnormal imaging findings. The minigene splicing result indicated that the c.1390-12T > G did not affect the splicing of CLCN4 mRNA. In vitro experiments showed that the mutant protein level and localization of mutant protein are similar to the wild type.
Conclusion: The study identified six probands with CLCN4 gene variants associated with X-linked ID. It expanded the gene and phenotype spectrum of CLCN4 variants. The bioinformatic analysis supported the pathogenicity of CLCN4 variants. However, these CLCN4 gene variants did not affect the ClC-4 expression levels and protein location, consistent with previous studies. Further investigations are necessary to investigate the pathogenetic mechanism.
Keywords: CLCN4 gene; MRXSRC; intellectual disability; missense variant; splice site variant.
Copyright © 2023 Li, Zhang, Liang, Zhu, Zheng, Zhou, Wang and Zhao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Palmer EE, Stuhlmann T, Weinert S, Haan E, van Esch H, Holvoet M, et al. . De novo and inherited mutations in the X-linked gene CLCN4 are associated with syndromic intellectual disability and behavior and seizure disorders in males and females. Mol Psychiatry. (2018) 23:222–30. 10.1038/mp.2016.135 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

