This is a preprint.
Radial glia promote microglial development through integrin αVβ8 -TGFβ1 signaling
- PMID: 37790363
- PMCID: PMC10542141
- DOI: 10.1101/2023.07.13.548459
Radial glia promote microglial development through integrin αVβ8 -TGFβ1 signaling
Update in
-
Radial glia integrin avb8 regulates cell autonomous microglial TGFβ1 signaling that is necessary for microglial identity.Nat Commun. 2025 Mar 22;16(1):2840. doi: 10.1038/s41467-025-57684-y. Nat Commun. 2025. PMID: 40121230 Free PMC article.
Abstract
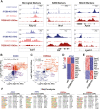
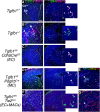
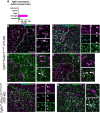
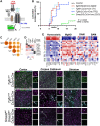
Microglia diversity emerges from interactions between intrinsic genetic programs and environment-derived signals, but how these processes unfold and interact in the developing brain remains unclear. Here, we show that radial glia-expressed integrin beta 8 (ITGB8) expressed in radial glia progenitors activates microglia-expressed TGFβ1, permitting microglial development. Domain-restricted deletion of Itgb8 in these progenitors establishes complementary regions with developmentally arrested "dysmature" microglia that persist into adulthood. In the absence of autocrine TGFβ1 signaling, we find that microglia adopt a similar dysmature phenotype, leading to neuromotor symptoms almost identical to Itgb8 mutant mice. In contrast, microglia lacking the TGFβ signal transducers Smad2 and Smad3 have a less polarized dysmature phenotype and correspondingly less severe neuromotor dysfunction. Finally, we show that non-canonical (Smad-independent) signaling partially suppresses disease and development associated gene expression, providing compelling evidence for the adoption of microglial developmental signaling pathways in the context of injury or disease.
Keywords: Microglia; TGFb signaling; development; differentiation; homeostasis; integrin αVβ8; neuroinflammation; radial glia; stem cells.
Conflict of interest statement
Declaration of interests D.S. and UCSF hold patents on the uses of antibodies that block the alphavbeta8 integrin. D.S. is a founder and owns stock in Pliant Therapeutics, is on the Scientific Review Board for Genentech and is on the Inflammation Scientific Review Board for Amgen.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
