This is a preprint.
Developmental progression of the nasopharyngeal microbiome during childhood and association with the lower airway microbiome
- PMID: 37790477
- PMCID: PMC10543049
- DOI: 10.1101/2023.09.18.23295747
Developmental progression of the nasopharyngeal microbiome during childhood and association with the lower airway microbiome
Update in
-
Upper and lower airway microbiota across infancy and childhood.Pediatr Res. 2025 Mar 12. doi: 10.1038/s41390-025-03942-0. Online ahead of print. Pediatr Res. 2025. PMID: 40075175
Abstract
Background: The upper (URT) and lower (LRT) respiratory tract feature distinct environments and responses affecting microbial colonization but investigating the relationship between them is technically challenging. We aimed to identify relationships between taxa colonizing the URT and LRT and explore their relationship with development during childhood.
Methods: We employed V4 16S rDNA sequencing to profile nasopharyngeal swabs and tracheal aspirates collected from 183 subjects between 20 weeks and 18 years of age. These samples were collected prior to elective procedures at the Children's Hospital of Philadelphia over the course of 20 weeks in 2020, from otherwise healthy subjects enrolled in a study investigating potential reservoirs of SARS-CoV-2.
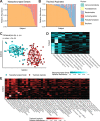
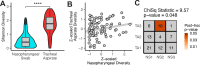
Findings: After extraction, sequencing, and quality control, we studied the remaining 124 nasopharyngeal swabs and 98 tracheal aspirates, including 85 subject-matched pairs of samples. V4 16S rDNA sequencing revealed that the nasopharynx is colonized by few, highly-abundant taxa, while the tracheal aspirates feature a diverse assembly of microbes. While no taxa co-occur in the URT and LRT of the same subject, clusters of microbiomes in the URT correlate with clusters of microbiomes in the LRT. The clusters identified in the URT correlate with subject age across childhood development.
Interpretations: The correlation between clusters of taxa across sites may suggest a mutual influence from either a third site, such as the oropharynx, or host-extrinsic, environmental features. The identification of a pattern of upper respiratory microbiota development across the first 18 years of life suggests that the patterns observed in early childhood may extend beyond the early life window.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- McCauley K. E. et al. Seasonal airway microbiome and transcriptome interactions promote childhood asthma exacerbations. J Allergy Clin Immunol 150, 204–213 (2022). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
