Modulation of Structure and Dynamics of Cardiac Troponin by Phosphorylation and Mutations Revealed by Molecular Dynamics Simulations
- PMID: 37791815
- PMCID: PMC10591477
- DOI: 10.1021/acs.jpcb.3c02337
Modulation of Structure and Dynamics of Cardiac Troponin by Phosphorylation and Mutations Revealed by Molecular Dynamics Simulations
Abstract
Adrenaline acts on β1 receptors in the heart muscle to enhance contractility, increase the heart rate, and increase the rate of relaxation (lusitropy) via activation of the cyclic AMP-dependent protein kinase, PKA. Phosphorylation of serines 22 and 23 in the N-terminal peptide of cardiac troponin I is responsible for lusitropy. Mutations associated with cardiomyopathy suppress the phosphorylation-dependent change. Key parts of troponin responsible for this modulatory system are disordered and cannot be resolved by conventional structural approaches. We performed all-atom molecular dynamics simulations (5 × 1.5
Conflict of interest statement
The authors declare no competing financial interest.
Figures

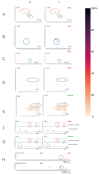
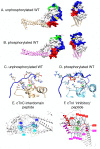
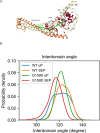
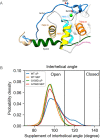
References
-
- Dong W. J.; Jayasundar J. J.; An J.; Xing J.; Cheung H. C. Effects of PKA phosphorylation of cardiac troponin I and strong crossbridge on conformational transitions of the N-domain of cardiac troponin C in regulated thin filaments. Biochemistry 2007, 46, 9752–9761. 10.1021/bi700574n. - DOI - PMC - PubMed
-
- Little S. C.; Biesiadecki B. J.; Kilic A.; Higgins R. S. D.; Janssen P. M. L.; Davis J. P. The rates of Ca2+ dissociation and cross-bridge detachment from ventricular myofibrils as reported by a fluorescent cardiac Troponin C. J. Biol. Chem. 2012, 287, 27930–27940. 10.1074/jbc.M111.337295. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

