Common scab disease: structural basis of elicitor recognition in pathogenic Streptomyces species
- PMID: 37791952
- PMCID: PMC10714786
- DOI: 10.1128/spectrum.01975-23
Common scab disease: structural basis of elicitor recognition in pathogenic Streptomyces species
Abstract
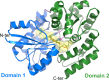
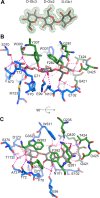
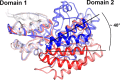
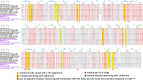
Common scab is a disease caused by a few Streptomyces species that affects important root and tuber crops including potato, beet, radish, and parsnip, resulting in major economic losses worldwide. In this work, we unveiled the molecular basis of host recognition by these pathogens by solving the structure of the sugar-binding protein CebE of Streptomyces scabiei in complex with cellotriose, the main elicitor of the pathogenic lifestyle of these bacteria. We further revealed that the signaling pathway from CebE-mediated transport of cellotriose is conserved in all pathogenic species except Streptomyces ipomoeae, which causes soft rot disease in sweet potatoes. Our work also provides the structural basis of the uptake of cellobiose and cellotriose in saprophytic Streptomyces species, the first step activating the expression of the enzymatic system degrading the most abundant polysaccharide on earth, cellulose.
Keywords: Streptomyces; carbohydrate metabolism; elicitor binding; host-pathogen interaction; ligand-protein interaction; plant pathogens; sugar transport.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Book AJ, Lewin GR, McDonald BR, Takasuka TE, Wendt-Pienkowski E, Doering DT, Suh S, Raffa KF, Fox BG, Currie CR, Hillis DM. 2016. Evolution of high cellulolytic activity in symbiotic Streptomyces through selection of expanded gene content and coordinated gene expression. PLOS Biol 14:e1002475. doi: 10.1371/journal.pbio.1002475 - DOI - PMC - PubMed
-
- Book AJ, Lewin GR, McDonald BR, Takasuka TE, Doering DT, Adams AS, Blodgett JAV, Clardy J, Raffa KF, Fox BG, Currie CR. 2014. Cellulolytic Streptomyces strains associated with herbivorous insects share a phylogenetically linked capacity to degrade lignocellulose. Appl Environ Microbiol 80:4692–4701. doi: 10.1128/AEM.01133-14 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CDR/OL J.0158.21/Fonds De La Recherche Scientifique - FNRS (FNRS)
- PDR/T.0121.22/Fonds De La Recherche Scientifique - FNRS (FNRS)
- FRIA 1.E.116.21/FNRS | Fonds pour la Formation à la Recherche dans l'Industrie et dans l'Agriculture (FRIA)
- 1.A250.13/Fonds De La Recherche Scientifique - FNRS (FNRS)
- 1.A618.18/Fonds De La Recherche Scientifique - FNRS (FNRS)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

