A Natural Language Processing Model for COVID-19 Detection Based on Dutch General Practice Electronic Health Records by Using Bidirectional Encoder Representations From Transformers: Development and Validation Study
- PMID: 37792444
- PMCID: PMC10563863
- DOI: 10.2196/49944
A Natural Language Processing Model for COVID-19 Detection Based on Dutch General Practice Electronic Health Records by Using Bidirectional Encoder Representations From Transformers: Development and Validation Study
Abstract
Background: Natural language processing (NLP) models such as bidirectional encoder representations from transformers (BERT) hold promise in revolutionizing disease identification from electronic health records (EHRs) by potentially enhancing efficiency and accuracy. However, their practical application in practice settings demands a comprehensive and multidisciplinary approach to development and validation. The COVID-19 pandemic highlighted challenges in disease identification due to limited testing availability and challenges in handling unstructured data. In the Netherlands, where general practitioners (GPs) serve as the first point of contact for health care, EHRs generated by these primary care providers contain a wealth of potentially valuable information. Nonetheless, the unstructured nature of free-text entries in EHRs poses challenges in identifying trends, detecting disease outbreaks, or accurately pinpointing COVID-19 cases.
Objective: This study aims to develop and validate a BERT model for detecting COVID-19 consultations in general practice EHRs in the Netherlands.
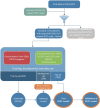
Methods: The BERT model was initially pretrained on Dutch language data and fine-tuned using a comprehensive EHR data set comprising confirmed COVID-19 GP consultations and non-COVID-19-related consultations. The data set was partitioned into a training and development set, and the model's performance was evaluated on an independent test set that served as the primary measure of its effectiveness in COVID-19 detection. To validate the final model, its performance was assessed through 3 approaches. First, external validation was applied on an EHR data set from a different geographic region in the Netherlands. Second, validation was conducted using results of polymerase chain reaction (PCR) test data obtained from municipal health services. Lastly, correlation between predicted outcomes and COVID-19-related hospitalizations in the Netherlands was assessed, encompassing the period around the outbreak of the pandemic in the Netherlands, that is, the period before widespread testing.
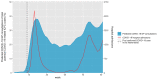
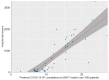
Results: The model development used 300,359 GP consultations. We developed a highly accurate model for COVID-19 consultations (accuracy 0.97, F1-score 0.90, precision 0.85, recall 0.85, specificity 0.99). External validations showed comparable high performance. Validation on PCR test data showed high recall but low precision and specificity. Validation using hospital data showed significant correlation between COVID-19 predictions of the model and COVID-19-related hospitalizations (F1-score 96.8; P<.001; R2=0.69). Most importantly, the model was able to predict COVID-19 cases weeks before the first confirmed case in the Netherlands.
Conclusions: The developed BERT model was able to accurately identify COVID-19 cases among GP consultations even preceding confirmed cases. The validated efficacy of our BERT model highlights the potential of NLP models to identify disease outbreaks early, exemplifying the power of multidisciplinary efforts in harnessing technology for disease identification. Moreover, the implications of this study extend beyond COVID-19 and offer a blueprint for the early recognition of various illnesses, revealing that such models could revolutionize disease surveillance.
Keywords: BERT model; COVID-19; EHR; NLP; disease identification; electronic health records; model development; multidisciplinary; natural language processing; prediction; primary care; public health.
©Maarten Homburg, Eline Meijer, Matthijs Berends, Thijmen Kupers, Tim Olde Hartman, Jean Muris, Evelien de Schepper, Premysl Velek, Jeroen Kuiper, Marjolein Berger, Lilian Peters. Originally published in the Journal of Medical Internet Research (https://www.jmir.org), 04.10.2023.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures
Similar articles
-
Multifaceted Natural Language Processing Task-Based Evaluation of Bidirectional Encoder Representations From Transformers Models for Bilingual (Korean and English) Clinical Notes: Algorithm Development and Validation.JMIR Med Inform. 2024 Oct 30;12:e52897. doi: 10.2196/52897. JMIR Med Inform. 2024. PMID: 39475725 Free PMC article.
-
Identifying Disinformation on the Extended Impacts of COVID-19: Methodological Investigation Using a Fuzzy Ranking Ensemble of Natural Language Processing Models.J Med Internet Res. 2025 May 21;27:e73601. doi: 10.2196/73601. J Med Internet Res. 2025. PMID: 40397945 Free PMC article.
-
Disease Concept-Embedding Based on the Self-Supervised Method for Medical Information Extraction from Electronic Health Records and Disease Retrieval: Algorithm Development and Validation Study.J Med Internet Res. 2021 Jan 27;23(1):e25113. doi: 10.2196/25113. J Med Internet Res. 2021. PMID: 33502324 Free PMC article.
-
Leveraging Natural Language Processing and Machine Learning Methods for Adverse Drug Event Detection in Electronic Health/Medical Records: A Scoping Review.Drug Saf. 2025 Apr;48(4):321-337. doi: 10.1007/s40264-024-01505-6. Epub 2025 Jan 9. Drug Saf. 2025. PMID: 39786481 Free PMC article.
-
The Growing Impact of Natural Language Processing in Healthcare and Public Health.Inquiry. 2024 Jan-Dec;61:469580241290095. doi: 10.1177/00469580241290095. Inquiry. 2024. PMID: 39396164 Free PMC article. Review.
Cited by
-
Generative artificial intelligence for general practice; new potential ahead, but are we ready?Eur J Gen Pract. 2025 Dec;31(1):2511645. doi: 10.1080/13814788.2025.2511645. Epub 2025 Jun 6. Eur J Gen Pract. 2025. PMID: 40478782 Free PMC article.
-
Hasselt Corona Impact Study: Impact of COVID-19 on healthcare seeking in a small Dutch town.NPJ Prim Care Respir Med. 2025 Apr 6;35(1):21. doi: 10.1038/s41533-025-00426-w. NPJ Prim Care Respir Med. 2025. PMID: 40188237 Free PMC article.
-
Implications of Data Extraction and Processing of Electronic Health Records for Epidemiological Research: Observational Study.J Med Internet Res. 2025 Jun 11;27:e64628. doi: 10.2196/64628. J Med Internet Res. 2025. PMID: 40498913 Free PMC article.
-
Year 2023 in Biomedical Natural Language Processing: a Tribute to Large Language Models and Generative AI.Yearb Med Inform. 2024 Aug;33(1):241-248. doi: 10.1055/s-0044-1800751. Epub 2025 Apr 8. Yearb Med Inform. 2024. PMID: 40199311 Free PMC article.
-
Integrating machine learning and artificial intelligence in life-course epidemiology: pathways to innovative public health solutions.BMC Med. 2024 Sep 2;22(1):354. doi: 10.1186/s12916-024-03566-x. BMC Med. 2024. PMID: 39218895 Free PMC article. Review.
References
-
- Jiang F, Jiang Y, Zhi H, Dong Y, Li H, Ma S, Wang Yilong, Dong Qiang, Shen Haipeng, Wang Yongjun. Artificial intelligence in healthcare: past, present and future. Stroke Vasc Neurol. 2017 Dec;2(4):230–243. doi: 10.1136/svn-2017-000101. https://svn.bmj.com/lookup/pmidlookup?view=long&pmid=29507784 svn-2017-000101 - DOI - PMC - PubMed
-
- Shimizu H, Nakayama K. Artificial intelligence in oncology. Cancer Sci. 2020 May;111(5):1452–1460. doi: 10.1111/cas.14377. https://europepmc.org/abstract/MED/32133724 - DOI - PMC - PubMed
-
- Nadkarni P, Ohno-Machado L, Chapman W. Natural language processing: an introduction. J Am Med Inform Assoc. 2011;18(5):544–51. doi: 10.1136/amiajnl-2011-000464. https://europepmc.org/abstract/MED/21846786 amiajnl-2011-000464 - DOI - PMC - PubMed
-
- Devlin J, Chang M, Lee K, Toutanova K. BERT: Pre-training of deep bidirectional transformers for language understanding. arXiv. Preprint posted online on October 11, 2018. doi: 10.48550/arXiv.1810.04805. https://arxiv.org/abs/1810.04805 - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

