O antigen biogenesis sensitises Escherichia coli K-12 to bile salts, providing a plausible explanation for its evolutionary loss
- PMID: 37792901
- PMCID: PMC10578602
- DOI: 10.1371/journal.pgen.1010996
O antigen biogenesis sensitises Escherichia coli K-12 to bile salts, providing a plausible explanation for its evolutionary loss
Abstract
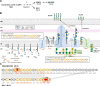
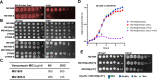
Escherichia coli K-12 is a model organism for bacteriology and has served as a workhorse for molecular biology and biochemistry for over a century since its first isolation in 1922. However, Escherichia coli K-12 strains are phenotypically devoid of an O antigen (OAg) since early reports in the scientific literature. Recent studies have reported the presence of independent mutations that abolish OAg repeating-unit (RU) biogenesis in E. coli K-12 strains from the same original source, suggesting unknown evolutionary forces have selected for inactivation of OAg biogenesis during the early propagation of K-12. Here, we show for the first time that restoration of OAg in E. coli K-12 strain MG1655 synergistically sensitises bacteria to vancomycin with bile salts (VBS). Suppressor mutants surviving lethal doses of VBS primarily contained disruptions in OAg biogenesis. We present data supporting a model where the transient presence and accumulation of lipid-linked OAg intermediates in the periplasmic leaflet of the inner membrane interfere with peptidoglycan sacculus biosynthesis, causing growth defects that are synergistically enhanced by bile salts. Lastly, we demonstrate that continuous bile salt exposure of OAg-producing MG1655 in the laboratory, can recreate a scenario where OAg disruption is selected for as an evolutionary fitness benefit. Our work thus provides a plausible explanation for the long-held mystery of the selective pressure that may have led to the loss of OAg biogenesis in E. coli K-12; this opens new avenues for exploring long-standing questions on the intricate network coordinating the synthesis of different cell envelope components in Gram-negative bacteria.
Copyright: © 2023 Qin et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors declare no competing interest exists.
Figures



References
-
- Bachmann bJ. Derivations and genotypes of some mutant derivatives of Escherichina coli K-12. In: Neidhardt FC, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. 2. 2 ed. New York: ASM Press; 1996.
-
- Clowes R. C. WH. Experiments in microbial genetics. New York: John Wiley & Sons Inc; 1968.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

