In-Cell Dynamics: The Next Focus of All-Atom Simulations
- PMID: 37793083
- PMCID: PMC10874638
- DOI: 10.1021/acs.jpcb.3c05166
In-Cell Dynamics: The Next Focus of All-Atom Simulations
Abstract
The cell is a crowded space where large biomolecules and metabolites are in continuous motion. Great strides have been made in in vitro studies of protein dynamics, folding, and protein-protein interactions, and much new data are emerging of how they differ in the cell. In this Perspective, we highlight the current progress in atomistic modeling of in-cell environments, both bacteria and mammals, with emphasis on classical all-atom molecular dynamics simulations. These simulations have been recently used to capture and characterize functional and non-functional protein-protein interactions, protein folding dynamics of small proteins with varied topologies, and dynamics of metabolites. We further discuss the challenges and efforts for updating modern force fields critical to the progress of cellular environment simulations. We also briefly summarize developments in relevant state-of-the-art experimental techniques. As computational and experimental methodologies continue to progress and produce more directly comparable data, we are poised to capture the complex atomistic picture of the cell.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

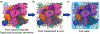
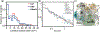
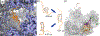
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

