The pseudotorsional space of RNA
- PMID: 37793790
- PMCID: PMC10653382
- DOI: 10.1261/rna.079821.123
The pseudotorsional space of RNA
Abstract
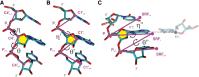
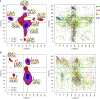
The characterization of the conformational landscape of the RNA backbone is rather complex due to the ability of RNA to assume a large variety of conformations. These backbone conformations can be depicted by pseudotorsional angles linking RNA backbone atoms, from which Ramachandran-like plots can be built. We explore here different definitions of these pseudotorsional angles, finding that the most accurate ones are the traditional η (eta) and θ (theta) angles, which represent the relative position of RNA backbone atoms P and C4'. We explore the distribution of η - θ in known experimental structures, comparing the pseudotorsional space generated with structures determined exclusively by one experimental technique. We found that the complete picture only appears when combining data from different sources. The maps provide a quite comprehensive representation of the RNA accessible space, which can be used in RNA-structural predictions. Finally, our results highlight that protein interactions lead to significant changes in the population of the η - θ space, pointing toward the role of induced-fit mechanisms in protein-RNA recognition.
Keywords: bioinformatics; computational chemistry; conformations; data mining; structure.
© 2023 Grille et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures





References
-
- Dans PD, Gallego D, Balaceanu A, Darré L, Gómez H, Orozco M. 2019. Modeling, simulations, and bioinformatics at the service of RNA structure. Chem 5: 51–73. 10.1016/j.chempr.2018.09.015 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
