TET2 lesions enhance the aggressiveness of CEBPA-mutant acute myeloid leukemia by rebalancing GATA2 expression
- PMID: 37794021
- PMCID: PMC10550934
- DOI: 10.1038/s41467-023-41927-x
TET2 lesions enhance the aggressiveness of CEBPA-mutant acute myeloid leukemia by rebalancing GATA2 expression
Abstract
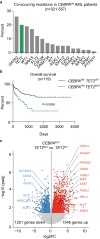
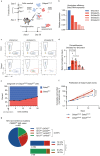
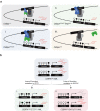
The myeloid transcription factor CEBPA is recurrently biallelically mutated (i.e., double mutated; CEBPADM) in acute myeloid leukemia (AML) with a combination of hypermorphic N-terminal mutations (CEBPANT), promoting expression of the leukemia-associated p30 isoform, and amorphic C-terminal mutations. The most frequently co-mutated genes in CEBPADM AML are GATA2 and TET2, however the molecular mechanisms underlying this co-mutational spectrum are incomplete. By combining transcriptomic and epigenomic analyses of CEBPA-TET2 co-mutated patients with models thereof, we identify GATA2 as a conserved target of the CEBPA-TET2 mutational axis, providing a rationale for the mutational spectra in CEBPADM AML. Elevated CEBPA levels, driven by CEBPANT, mediate recruitment of TET2 to the Gata2 distal hematopoietic enhancer thereby increasing Gata2 expression. Concurrent loss of TET2 in CEBPADM AML induces a competitive advantage by increasing Gata2 promoter methylation, thereby rebalancing GATA2 levels. Of clinical relevance, demethylating treatment of Cebpa-Tet2 co-mutated AML restores Gata2 levels and prolongs disease latency.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

