Large-scale plasma proteomics comparisons through genetics and disease associations
- PMID: 37794188
- PMCID: PMC10567571
- DOI: 10.1038/s41586-023-06563-x
Large-scale plasma proteomics comparisons through genetics and disease associations
Erratum in
-
Author Correction: Large-scale plasma proteomics comparisons through genetics and disease associations.Nature. 2024 Jun;630(8015):E3. doi: 10.1038/s41586-024-07549-z. Nature. 2024. PMID: 38778117 Free PMC article. No abstract available.
Abstract
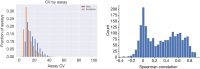
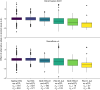
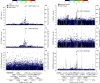
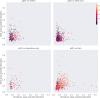
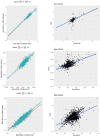
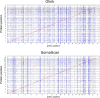
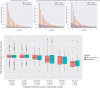
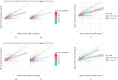
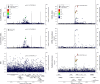
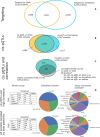
High-throughput proteomics platforms measuring thousands of proteins in plasma combined with genomic and phenotypic information have the power to bridge the gap between the genome and diseases. Here we performed association studies of Olink Explore 3072 data generated by the UK Biobank Pharma Proteomics Project1 on plasma samples from more than 50,000 UK Biobank participants with phenotypic and genotypic data, stratifying on British or Irish, African and South Asian ancestries. We compared the results with those of a SomaScan v4 study on plasma from 36,000 Icelandic people2, for 1,514 of whom Olink data were also available. We found modest correlation between the two platforms. Although cis protein quantitative trait loci were detected for a similar absolute number of assays on the two platforms (2,101 on Olink versus 2,120 on SomaScan), the proportion of assays with such supporting evidence for assay performance was higher on the Olink platform (72% versus 43%). A considerable number of proteins had genomic associations that differed between the platforms. We provide examples where differences between platforms may influence conclusions drawn from the integration of protein levels with the study of diseases. We demonstrate how leveraging the diverse ancestries of participants in the UK Biobank helps to detect novel associations and refine genomic location. Our results show the value of the information provided by the two most commonly used high-throughput proteomics platforms and demonstrate the differences between them that at times provides useful complementarity.
© 2023. The Author(s).
Conflict of interest statement
All authors are employees of deCODE Genetics, a wholly-owned subsidiary of Amgen.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

