Multivariate GWAS of Alzheimer's disease CSF biomarker profiles implies GRIN2D in synaptic functioning
- PMID: 37794492
- PMCID: PMC10548686
- DOI: 10.1186/s13073-023-01233-z
Multivariate GWAS of Alzheimer's disease CSF biomarker profiles implies GRIN2D in synaptic functioning
Abstract
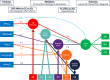
Background: Genome-wide association studies (GWAS) of Alzheimer's disease (AD) have identified several risk loci, but many remain unknown. Cerebrospinal fluid (CSF) biomarkers may aid in gene discovery and we previously demonstrated that six CSF biomarkers (β-amyloid, total/phosphorylated tau, NfL, YKL-40, and neurogranin) cluster into five principal components (PC), each representing statistically independent biological processes. Here, we aimed to (1) identify common genetic variants associated with these CSF profiles, (2) assess the role of associated variants in AD pathophysiology, and (3) explore potential sex differences.
Methods: We performed GWAS for each of the five biomarker PCs in two multi-center studies (EMIF-AD and ADNI). In total, 973 participants (n = 205 controls, n = 546 mild cognitive impairment, n = 222 AD) were analyzed for 7,433,949 common SNPs and 19,511 protein-coding genes. Structural equation models tested whether biomarker PCs mediate genetic risk effects on AD, and stratified and interaction models probed for sex-specific effects.
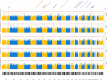
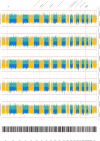
Results: Five loci showed genome-wide significant association with CSF profiles, two were novel (rs145791381 [inflammation] and GRIN2D [synaptic functioning]) and three were previously described (APOE, TMEM106B, and CHI3L1). Follow-up analyses of the two novel signals in independent datasets only supported the GRIN2D locus, which contains several functionally interesting candidate genes. Mediation tests indicated that variants in APOE are associated with AD status via processes related to amyloid and tau pathology, while markers in TMEM106B and CHI3L1 are associated with AD only via neuronal injury/inflammation. Additionally, seven loci showed sex-specific associations with AD biomarkers.
Conclusions: These results suggest that pathway and sex-specific analyses can improve our understanding of AD genetics and may contribute to precision medicine.
Keywords: Alzheimer’s disease; Biomarkers; Cerebrospinal fluid (CSF); Dementia; Genome-wide association study (GWAS); Mediation; Multivariate analysis; Principal component analysis; Structural equation modeling.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
FB is on the steering committee or iDMC member for Biogen, Merck, Roche, EISAI, and Prothena. consultant for Roche, Biogen, Merck, IXICO, Jansen, Combinostics, has research agreements with Merck, Biogen, GE Healthcare, Roche, and is co-founder and shareholder of Queen Square Analytics LTD; HZ has served at scientific advisory boards and/or as a consultant for Abbvie, Alector, ALZPath, Annexon, Apellis, Artery Therapeutics, AZTherapies, CogRx, Denali, Eisai, Nervgen, Novo Nordisk, Passage Bio, Pinteon Therapeutics, Red Abbey Labs, reMYND, Roche, Samumed, Siemens Healthineers, Triplet Therapeutics, and Wave, has given lectures in symposia sponsored by Cellectricon, Fujirebio, Alzecure, Biogen, and Roche, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program (outside submitted work). AL is on the editorial board of Neurology and Brain Communications, has served at scientific advisory boards from Fujirebio-Europe, Nutricia, Roche-Genentech, Biogen, Grifols, and Roche Diagnostics, and has filed a patent application of synaptic markers in neurodegenerative diseases. D.A. participated in advisory boards from Fujirebio-Europe and Roche Diagnostics and received speaker honoraria from Fujirebio-Europe, Roche Diagnostics, Nutricia, Krka Farmacéutica S.L., Zambon S.A.U., and Esteve Pharmaceuticals S.A. D.A. declares a filed patent application (WO2019175379 A1 Markers of synaptopathy in neurodegenerative disease). SL is currently an employee of Janssen Medical Ltd (UK), a co-founder of Akrivia Health Ltd (UK) and within the past 5 years has filed patents related to biomarkers unrelated to the current work and advised or given lectures for Merck, Optum Labs, and Eisai as well as having received grant funding from multiple companies as part of EU IMI programs and from Astra Zeneca. JP received consultation honoraria from Nestle Institute of Health Sciences, Ono Pharma, OM Pharma, and Fujirebio, unrelated to the submitted work. SE has served on scientific advisory boards for Biogen, Danone, icometrix, Novartis, Nutricia, and Roche and received unrestricted research grants from Janssen Pharmaceutica and ADx Neurosciences (paid to institution). JR was an employee at GSK and currently an employee at the MSD London Discovery Centre, U.K. CC has received research support from: GSK and EISAI. CC is a member of the advisory board of Vivid Genomics and Circular Genomics and owns stocks. The remaining authors declare that they have no competing interests.
Figures

References
-
- Sims R, Hill M, Williams J. The multiplex model of the genetics of Alzheimer’s disease. Nat Neurosci. 2020;23:311–322. - PubMed
-
- Olsson B, Lautner R, Andreasson U, Öhrfelt A, Portelius E, Bjerke M, Hölttä M, Rosén C, Olsson C, Strobel G, et al. CSF and blood biomarkers for the diagnosis of Alzheimer’s disease: a systematic review and meta-analysis. Lancet Neurol. 2016;15:673–684. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG018023/AG/NIA NIH HHS/United States
- 101039672/ERC_/European Research Council/International
- RF1 AG053303/AG/NIA NIH HHS/United States
- P30 AG019610/AG/NIA NIH HHS/United States
- U01 AG058922/AG/NIA NIH HHS/United States
- U24 NS072026/NS/NINDS NIH HHS/United States
- U01 AG006786/AG/NIA NIH HHS/United States
- RF1 AG057440/AG/NIA NIH HHS/United States
- P50 AG016574/AG/NIA NIH HHS/United States
- R01 AG064877/AG/NIA NIH HHS/United States
- R01 AG032990/AG/NIA NIH HHS/United States
- R01 AG044546/AG/NIA NIH HHS/United States
- DH_/Department of Health/United Kingdom
- P01 AG003991/AG/NIA NIH HHS/United States
- U01 AG046139/AG/NIA NIH HHS/United States
- R01 AG039495/AG/NIA NIH HHS/United States
- U01 AG061356/AG/NIA NIH HHS/United States
- P01 AG017216/AG/NIA NIH HHS/United States
- RF1 AG058501/AG/NIA NIH HHS/United States
- U01 AG024904/AG/NIA NIH HHS/United States
- R01 AG015819/AG/NIA NIH HHS/United States
- U01 AG046170/AG/NIA NIH HHS/United States
- R01 NS080820/NS/NINDS NIH HHS/United States
- U24 AG061340/AG/NIA NIH HHS/United States
- P30 AG072975/AG/NIA NIH HHS/United States
- RF1 AG044546/AG/NIA NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- U01 AG046152/AG/NIA NIH HHS/United States
- R01 AG036836/AG/NIA NIH HHS/United States
- R01 AG017917/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

