The Clinical Utility of Genetic Testing in the Diagnosis and Management of Adults with Chronic Kidney Disease
- PMID: 37794564
- PMCID: PMC10703084
- DOI: 10.1681/ASN.0000000000000249
The Clinical Utility of Genetic Testing in the Diagnosis and Management of Adults with Chronic Kidney Disease
Abstract
Significance statement: Accurate diagnosis of a patient's underlying cause of CKD can influence management and ultimately overall health. The single-arm, interventional, prospective Renasight Clinical Application, Review, and Evaluation study assessed the utility of genetic testing with a 385 gene kidney disease panel on the diagnosis and management of 1623 patients with CKD. Among 20.8% of patients who had positive genetic findings, half resulted in a new or reclassified diagnosis. In addition, a change in management because of genetic testing was reported for 90.7% of patients with positive findings, including treatment changes in 32.9%. These findings demonstrate that genetic testing has a significant effect on both CKD diagnosis and management.
Background: Genetic testing in CKD has recently been shown to have diagnostic utility with many predicted implications for clinical management, but its effect on management has not been prospectively evaluated.
Methods: Renasight Clinical Application, Review, and Evaluation RenaCARE (ClinicalTrials.gov NCT05846113 ) is a single-arm, interventional, prospective, multicenter study that evaluated the utility of genetic testing with a broad, 385 gene panel (the Renasight TM test) on the diagnosis and management of adult patients with CKD recruited from 31 US-based community and academic medical centers. Patient medical history and clinical CKD diagnosis were collected at enrollment. Physician responses to questionnaires regarding patient disease categorization and management were collected before genetic testing and 1 month after the return of test results. Changes in CKD diagnosis and management after genetic testing were assessed.
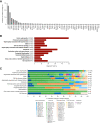
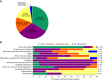
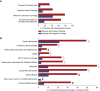
Results: Of 1623 patients with CKD in 13 predefined clinical disease categories (ages, 18-96; median, 55 years), 20.8% ( n =338) had positive genetic findings spanning 54 genes. Positive genetic findings provided a new diagnosis or reclassified a prior diagnosis in 48.8% of those patients. Physicians reported that genetic results altered the management of 90.7% of patients with a positive genetic finding, including changes in treatment plan, which were reported in 32.9% of these patients.
Conclusions: Genetic testing with a CKD-focused 385 gene panel substantially refined clinical diagnoses and had widespread implications for clinical management, including appropriate treatment strategies. These data support the utility of broader integration of panels of genetic tests into the clinical care paradigm for patients with CKD.
Clinical trial registry name and registration number: ClinicalTrials.gov, NCT05846113 .
Copyright © 2023 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the American Society of Nephrology.
Conflict of interest statement
S. Bhorade reports Employer: Natera and Veracyte; Ownership Interest: Natera and Veracyte; and Patents or Royalties: Veracyte. S. Bhorade, M.S. Bloom, D. Clark, Z.P. Demko, S. Jandeska, K. Marshall, S. Punj, H. Tabriziani, M. Westemeyer, and Z. Zhang are employees of Natera, Inc. and own or have the option to own stock. V. Broumand reports Consultancy: Bayer, Outset Medical, and Vifor Pharma; Ownership Interest: ADR, Ardelyx, Outset Medical, Precigen, Renalytx, and US Renal Care; Research Funding: AMAG Pharmaceuticals, Astra Zenca, Fibrogen, Kind Pharmaceuticals, and Pathalys Pharma; Honoraria: Bayer, Outset Medical, and Vifor Pharma; Advisory or Leadership Role: Chief Medical Officer, Christus Santa Rosa Westover Hills Hospital, and Health System Dialysis Medical Director; and Speakers Bureau: Bayer, Outset Medical, Natera, and Vifor Pharma. F.T. Chebib has received research and educational fellowship grant funding from Otsuka Pharmaceuticals. F.T. Chebib also reports Patents or Royalties: Patent no US20200368191A1; and Advisory or Leadership Role: PKD Foundation—Chair of Education Advisory Panel. N.K. Dahl reports Consultancy: Otsuka Pharmaceuticals; Research Funding: PI for clinical trials sponsored by Reata, and Vertex; Honoraria: Natera and Otsuka Pharmaceuticals; Advisory or Leadership Role: Natera Scientific Advisory Board and PKD Foundation; Speakers Bureau: on the unbranded speakers bureau for Otsuka until December 2022; and Other Interests or Relationships: Medical Advisory Board and NKF NE Chapter. R.A. Fatica reports Research Funding: Natera and Reata. A.G. Gharavi receives a research grant from Natera and has served on advisory boards for Natera through a service agreement with Columbia University. A.G. Gharavi has served on advisory boards for Actio Biosciences, Alnylam, Novartis, and Travere and has stock options for Actio Biosciences. A.G. Gharavi also reports Consultancy: Actio Biosciences, Alnylam, Novartis, and Travere; Research Funding: Renal Research Institute; Honoraria: Alnylam and Sanofi; and Advisory or Leadership Role: Editorial board:
Figures

References
-
- Global Health Estimates 2020. Deaths by Cause, Age, Sex, by Country and by Region, 2000-2019. Organization WH (Ed.); 2020.
-
- United States Renal Data System. 2022 USRDS Annual Data Report: Epidemiology of Kidney Disease in the United States. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD, 2022.
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

