SND1 binds SARS-CoV-2 negative-sense RNA and promotes viral RNA synthesis through NSP9
- PMID: 37794589
- PMCID: PMC10617981
- DOI: 10.1016/j.cell.2023.09.002
SND1 binds SARS-CoV-2 negative-sense RNA and promotes viral RNA synthesis through NSP9
Abstract
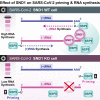
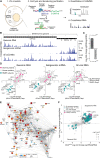
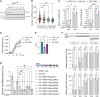
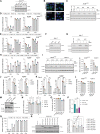
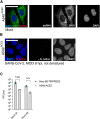
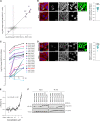
Regulation of viral RNA biogenesis is fundamental to productive SARS-CoV-2 infection. To characterize host RNA-binding proteins (RBPs) involved in this process, we biochemically identified proteins bound to genomic and subgenomic SARS-CoV-2 RNAs. We find that the host protein SND1 binds the 5' end of negative-sense viral RNA and is required for SARS-CoV-2 RNA synthesis. SND1-depleted cells form smaller replication organelles and display diminished virus growth kinetics. We discover that NSP9, a viral RBP and direct SND1 interaction partner, is covalently linked to the 5' ends of positive- and negative-sense RNAs produced during infection. These linkages occur at replication-transcription initiation sites, consistent with NSP9 priming viral RNA synthesis. Mechanistically, SND1 remodels NSP9 occupancy and alters the covalent linkage of NSP9 to initiating nucleotides in viral RNA. Our findings implicate NSP9 in the initiation of SARS-CoV-2 RNA synthesis and unravel an unsuspected role of a cellular protein in orchestrating viral RNA production.
Keywords: RNA binding proteins; RNA biology; RNA interactome; RNA virus; SARS-CoV-2; host factors; omics technologies; proteomics; systems biology; virus host interactions.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

