Interactions of Indoleamine 2,3-dioxygenase-expressing LAMP3+ dendritic cells with CD4+ regulatory T cells and CD8+ exhausted T cells: synergistically remodeling of the immunosuppressive microenvironment in cervical cancer and therapeutic implications
- PMID: 37794698
- PMCID: PMC10631485
- DOI: 10.1002/cac2.12486
Interactions of Indoleamine 2,3-dioxygenase-expressing LAMP3+ dendritic cells with CD4+ regulatory T cells and CD8+ exhausted T cells: synergistically remodeling of the immunosuppressive microenvironment in cervical cancer and therapeutic implications
Abstract
Background: Cervical cancer (CC) is the fourth most common cancer in women worldwide. Although immunotherapy has been applied in clinical practice, its therapeutic efficacy remains far from satisfactory, necessitating further investigation of the mechanism of CC immune remodeling and exploration of novel treatment targets. This study aimed to investigate the mechanism of CC immune remodeling and explore potential therapeutic targets.
Methods: We conducted single-cell RNA sequencing on a total of 17 clinical specimens, including normal cervical tissues, high-grade squamous intraepithelial lesions, and CC tissues. To validate our findings, we conducted multicolor immunohistochemical staining of CC tissues and constructed a subcutaneous tumorigenesis model in C57BL/6 mice using murine CC cell lines (TC1) to evaluate the effectiveness of combination therapy involving indoleamine 2,3-dioxygenase 1 (IDO1) inhibition and immune checkpoint blockade (ICB). We used the unpaired two-tailed Student's t-test, Mann-Whitney test, or Kruskal-Wallis test to compare continuous data between two groups and one-way ANOVA with Tukey's post hoc test to compare data between multiple groups.
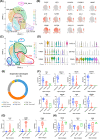
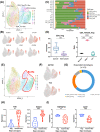
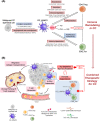
Results: Malignant cervical epithelial cells did not manifest noticeable signs of tumor escape, whereas lysosomal-associated membrane protein 3-positive (LAMP3+ ) dendritic cells (DCs) in a mature state with immunoregulatory roles were found to express IDO1 and affect tryptophan metabolism. These cells interacted with both tumor-reactive exhausted CD8+ T cells and CD4+ regulatory T cells, synergistically forming a vicious immunosuppressive cycle and mediating CC immune escape. Further validation through multicolor immunohistochemical staining showed co-localization of neoantigen-reactive T cells (CD3+ , CD4+ /CD8+ , and PD-1+ ) and LAMP3+ DCs (CD80+ and PD-L1+ ). Additionally, a combination of the IDO1 inhibitor with an ICB agent significantly reduced tumor volume in the mouse model of CC compared with an ICB agent alone.
Conclusions: Our study suggested that a combination treatment consisting of targeting IDO1 and ICB agent could improve the therapeutic efficacy of current CC immunotherapies. Additionally, our results provided crucial insights for designing drugs and conducting future clinical trials for CC.
Keywords: T cell; cervical cancer; dendritic cell; immune checkpoint blockade; immune escape; indoleamine 2,3-dioxygenase 1; single-cell analysis.
© 2023 The Authors. Cancer Communications published by John Wiley & Sons Australia, Ltd on behalf of SUN YAT-SEN UNIVERSITY CANCER CENTER.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. - PubMed
-
- Chung HC, Ros W, Delord JP, Perets R, Italiano A, Shapira‐Frommer R, et al. Efficacy and Safety of pembrolizumab in previously treated advanced cervical cancer: results from the Phase II KEYNOTE‐158 study. J Clin Oncol. 2019;37(17):1470–1478. - PubMed
-
- O'Malley DM, Oaknin A, Monk BJ, Selle F, Rojas C, Gladieff L, et al. Phase II study of the safety and efficacy of the anti‐PD‐1 antibody balstilimab in patients with recurrent and/or metastatic cervical cancer. Gynecol Oncol. 2021;163(2):274–280. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

