ARPC1B is a novel prognostic biomarker for kidney renal clear cell carcinoma and correlates with immune infiltration
- PMID: 37795220
- PMCID: PMC10546172
- DOI: 10.3389/fmolb.2023.1202524
ARPC1B is a novel prognostic biomarker for kidney renal clear cell carcinoma and correlates with immune infiltration
Abstract
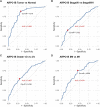
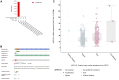
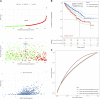
Background: Actin-related protein 2/3 complex subunit 1B (ARPC1B) is reported to be involved in tumorigenesis and progression. However, its role in kidney renal clear cell carcinoma (KIRC), correlation with tumor-infiltrating immune cells, and prognostic significance remain unclear. Methods: Data sets from the TCGA, GTEx, GEPIA, GEO, UALCAN, and CPTAC databases were extracted and analyzed to investigate the expression difference, prognosis, and clinicopathological features of ARPC1B. Single-sample Gene Set Enrichment Analysis (ssGSEA), CIBERSORT, and TISCH2 analysis were used to examine the relationship between ARPC1B expression and tumor immune infiltration in KIRC. The potential function of ARPC1B in KIRC was explored by GO functional annotation and KEGG pathway analysis. The TIDE algorithm was used to predict and analyze the relationship between ARPC1B expression and response to immune checkpoint blockade (ICB). The expression of ARPC1B was further validated by using quantitative real-time polymerase chain reaction (qRT-PCR) and immunohistochemistry (IHC). Results: The study showed that ARPC1B expression was an independent prognostic factor of KIRC, with high ARPC1B expression being associated with poor overall survival (OS). Enrichment of GO annotation and pathway analysis showed multiple immune-related functional pathways affected by ARPC1B such as regulation of immune effector process, inflammatory response regulation, antigen processing and presentation, asthma, autoimmune thyroid disease, graft versus host disease, intestinal immune network for IgA production, and type I diabetic mellitus. Moreover, ARPC1B expression positively correlated with infiltrating levels of myeloid-derived suppressor cells (MDSCs) and regulatory T cells (Tregs) in KIRC. Importantly, high ARPC1B expression predicted a low response to ICB in KIRC. Conclusion: This study indicates that ARPC1B expression is an independent prognostic biomarker for OS in KIRC patients. High ARPC1B expression is closely associated with MDSCs and Tregs infiltration. These findings suggest that ARPC1B may serve as a biomarker for prognosis and immune infiltration in KIRC, potentially aiding in the development of novel treatment strategies to improve the survival outcomes for KIRC patients.
Keywords: ARPC1B; Arp2/3 protein complex; immunotherapy; kidney renal clear cell carcinoma; tumor immune microenvironment.
Copyright © 2023 Tang, Qiao, Huang and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







Similar articles
-
Prognostic biomarker NEIL3 and its association with immune infiltration in renal clear cell carcinoma.Front Oncol. 2023 Feb 2;13:1073941. doi: 10.3389/fonc.2023.1073941. eCollection 2023. Front Oncol. 2023. PMID: 36816967 Free PMC article.
-
BAG3 as a novel prognostic biomarker in kidney renal clear cell carcinoma correlating with immune infiltrates.Eur J Med Res. 2024 Feb 1;29(1):93. doi: 10.1186/s40001-024-01687-w. Eur J Med Res. 2024. PMID: 38297320 Free PMC article.
-
Cuproptosis-related gene FDX1 as a prognostic biomarker for kidney renal clear cell carcinoma correlates with immune checkpoints and immune cell infiltration.Front Genet. 2023 Jan 23;14:1071694. doi: 10.3389/fgene.2023.1071694. eCollection 2023. Front Genet. 2023. PMID: 36755576 Free PMC article.
-
CD72, a new immune checkpoint molecule, is a novel prognostic biomarker for kidney renal clear cell carcinoma.Eur J Med Res. 2023 Nov 18;28(1):531. doi: 10.1186/s40001-023-01487-8. Eur J Med Res. 2023. PMID: 37980541 Free PMC article.
-
GRAMD1A Is a Biomarker of Kidney Renal Clear Cell Carcinoma and Is Associated with Immune Infiltration in the Tumour Microenvironment.Dis Markers. 2022 Jul 11;2022:5939021. doi: 10.1155/2022/5939021. eCollection 2022. Dis Markers. 2022. PMID: 35860689 Free PMC article.
Cited by
-
Potential role of P4HB in the tumor microenvironment and its clinical prognostic value: a comprehensive pan-cancer analysis and experimental validation with a focus on KIRC.Cancer Cell Int. 2025 Jan 3;25(1):1. doi: 10.1186/s12935-024-03575-z. Cancer Cell Int. 2025. PMID: 39754183 Free PMC article.
References
-
- Auzair L. B., Vincent-Chong V. K., Ghani W. M., Kallarakkal T. G., Ramanathan A., Lee C. E., et al. (2016). Caveolin 1 (Cav-1) and actin-related protein 2/3 complex, subunit 1B (ARPC1B) expressions as prognostic indicators for oral squamous cell carcinoma (OSCC). Eur. Arch. Otorhinolaryngol. 273 (7), 1885–1893. 10.1007/s00405-015-3703-9 - DOI - PubMed
-
- Bahadoram S., Davoodi M., Hassanzadeh S., Bahadoram M., Barahman M., Mafakher L. (2022). Renal cell carcinoma: an overview of the epidemiology, diagnosis, and treatment. G. Ital. Nefrol. 39 (3), 2022–vol3. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

