RAMP1 as a novel prognostic biomarker in pan-cancer and osteosarcoma
- PMID: 37796823
- PMCID: PMC10553254
- DOI: 10.1371/journal.pone.0292452
RAMP1 as a novel prognostic biomarker in pan-cancer and osteosarcoma
Abstract
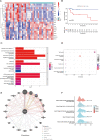
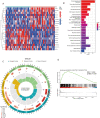
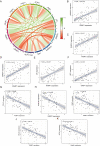
Receptor activity modifying protein 1 (RAMP1) facilitates the localization of the calcitonin-like receptor (CLR) to the plasma membrane, but its role in osteosarcoma (OS) remains unclear. We evaluated the RAMP1 expression and prognostic value across different cancers, studying tumor immune infiltration. The prognostic value was analyzed using the GSE39058 and TARGET datasets. Differential gene expression was evaluated. a protein-protein interaction network was constructed, and gene set enrichment analysis was performed. The function of RAMP1 in the tumor microenvironment was analyzed, and its expression in OS cell lines was validated using quantitative real-time PCR. High RAMP1 expression correlated with poor prognosis relative to low RAMP1 expression (p < 0.05). Low RAMP1 expression correlated with an abundance of CD4+ memory-activated T cells. whereas a high expression level correlated with a high proportion of gamma-delta T cells (γδ T cells). Differentially expressed genes from TARGET was enriched in olfactory transduction pathways (normalized enrichment scores [NES] = 1.6998, p < 0.0001). RAMP1 expression negatively correlated with CD44 expression but positively correlated with TNFSF9 expression. The RAMP1 gene is substantially expressed in OS cells compared to the normal osteoblast cell line hFOB1.19. Thus, RAMP1 may be a prognostic biomarker and potential therapeutic target in OS.
Copyright: © 2023 Xie et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Localization of calcitonin receptor-like receptor and receptor activity modifying protein 1 in enteric neurons, dorsal root ganglia, and the spinal cord of the rat.J Comp Neurol. 2005 Sep 26;490(3):239-55. doi: 10.1002/cne.20669. J Comp Neurol. 2005. PMID: 16082677
-
Post-endocytic sorting of calcitonin receptor-like receptor and receptor activity-modifying protein 1.J Biol Chem. 2007 Apr 20;282(16):12260-71. doi: 10.1074/jbc.M606338200. Epub 2007 Feb 19. J Biol Chem. 2007. PMID: 17310067
-
Activation of calcitonin receptor and calcitonin receptor-like receptor by membrane-anchored ligands.J Biol Chem. 2010 Jan 8;285(2):1075-80. doi: 10.1074/jbc.M109.020040. Epub 2009 Nov 10. J Biol Chem. 2010. PMID: 19903822 Free PMC article.
-
Calcitonin and Amylin Receptor Peptide Interaction Mechanisms: INSIGHTS INTO PEPTIDE-BINDING MODES AND ALLOSTERIC MODULATION OF THE CALCITONIN RECEPTOR BY RECEPTOR ACTIVITY-MODIFYING PROTEINS.J Biol Chem. 2016 Apr 15;291(16):8686-700. doi: 10.1074/jbc.M115.713628. Epub 2016 Feb 19. J Biol Chem. 2016. PMID: 26895962 Free PMC article.
-
Cerebellar distribution of calcitonin gene-related peptide (CGRP) and its receptor components calcitonin receptor-like receptor (CLR) and receptor activity modifying protein 1 (RAMP1) in rat.Mol Cell Neurosci. 2011 Jan;46(1):333-9. doi: 10.1016/j.mcn.2010.10.005. Epub 2010 Oct 30. Mol Cell Neurosci. 2011. PMID: 21040789
Cited by
-
Identification of crosstalk genes and immune characteristics between Alzheimer's disease and atherosclerosis.Front Immunol. 2024 Aug 12;15:1443464. doi: 10.3389/fimmu.2024.1443464. eCollection 2024. Front Immunol. 2024. PMID: 39188714 Free PMC article.
-
CD47 and IFT57 Are Colinear Genes That Are Highly Coexpressed in Most Cancers and Exhibit Parallel Cancer-Specific Correlations with Survival.Int J Mol Sci. 2024 Aug 17;25(16):8956. doi: 10.3390/ijms25168956. Int J Mol Sci. 2024. PMID: 39201643 Free PMC article.
-
Multi-cohort validation based on a novel prognostic signature of anoikis for predicting prognosis and immunotherapy response of esophageal squamous cell carcinoma.Front Oncol. 2025 Mar 17;15:1530035. doi: 10.3389/fonc.2025.1530035. eCollection 2025. Front Oncol. 2025. PMID: 40165896 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

