Chromosome-level assembly and annotation of the Xyrichtys novacula (Linnaeus, 1758) genome
- PMID: 37797305
- PMCID: PMC10590160
- DOI: 10.1093/dnares/dsad021
Chromosome-level assembly and annotation of the Xyrichtys novacula (Linnaeus, 1758) genome
Abstract
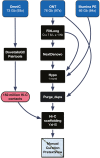
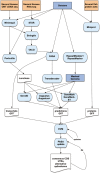
The pearly razorfish (Xyrichtys novacula), commonly known as raor in the Balearic Islands, is a wrasse within the family Labridae. This fish species has particular biological and socio-cultural characteristics making it an ideal model organism in the fields of behavioural ecology, molecular ecology and conservation biology. In this study, we present the first annotated chromosome-level assembly for this species. Sequencing involved a combination of long reads with Oxford Nanopore Technologies, Illumina paired-end short reads (2 × 151 bp), Hi-C and RNA-seq from different tissues. The nuclear genome assembly has a scaffold N50 of 34.33 Mb, a total assembly span of 775.53 Mb and 99.63% of the sequence assembled into 24 superscaffolds, consistent with its known karyotype. Quality metrics revealed a consensus accuracy (QV) of 42.92 and gene completeness > 98%. The genome annotation resulted in 26,690 protein-coding genes and 12,737 non-coding transcripts. The coding regions encoded 39,613 unique protein products, 93% of them with assigned function. Overall, the publication of the X. novacula's reference genome will broaden the scope and impact of genomic research conducted on this iconic and colourful species.
Keywords: Hi-C scaffolding; Labridae; Xyrichtys novacula; chromosome-level assembly; genome annotation.
© The Author(s) 2023. Published by Oxford University Press on behalf of Kazusa DNA Research Institute.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Complete mitochondrial genome of the pearly razorfish Xyrichtys novacula: phylogenetic analysis of its placement within the Labridae family.Mitochondrial DNA B Resour. 2020 Jan 16;5(1):644-645. doi: 10.1080/23802359.2019.1711226. Mitochondrial DNA B Resour. 2020. PMID: 33366684 Free PMC article.
-
Chromosome-Level Genome Assembly and Annotation of Corallium rubrum: A Mediterranean Coral Threatened by Overharvesting and Climate Change.Genome Biol Evol. 2025 Feb 3;17(2):evae253. doi: 10.1093/gbe/evae253. Genome Biol Evol. 2025. PMID: 39917963 Free PMC article.
-
Chromosome-level genome assembly of Lilford's wall lizard, Podarcis lilfordi (Günther, 1874) from the Balearic Islands (Spain).DNA Res. 2023 Jun 1;30(3):dsad008. doi: 10.1093/dnares/dsad008. DNA Res. 2023. PMID: 37137526 Free PMC article.
-
Finding Nemo: hybrid assembly with Oxford Nanopore and Illumina reads greatly improves the clownfish (Amphiprion ocellaris) genome assembly.Gigascience. 2018 Mar 1;7(3):1-6. doi: 10.1093/gigascience/gix137. Gigascience. 2018. PMID: 29342277 Free PMC article.
-
Chromosome-level genome assembly and annotation of the Antarctica whitefin plunderfish Pogonophryne albipinna.Sci Data. 2023 Dec 12;10(1):891. doi: 10.1038/s41597-023-02811-x. Sci Data. 2023. PMID: 38086886 Free PMC article.
Cited by
-
The BioGenome Portal: a web-based platform for biodiversity genomics data management.NAR Genom Bioinform. 2025 Mar 22;7(1):lqaf020. doi: 10.1093/nargab/lqaf020. eCollection 2025 Mar. NAR Genom Bioinform. 2025. PMID: 40124712 Free PMC article.
-
The Catalan initiative for the Earth BioGenome Project: contributing local data to global biodiversity genomics.NAR Genom Bioinform. 2024 Jul 17;6(3):lqae075. doi: 10.1093/nargab/lqae075. eCollection 2024 Sep. NAR Genom Bioinform. 2024. PMID: 39022326 Free PMC article.
-
Atlantic mackerel population structure does not support genetically distinct spawning components.Open Res Eur. 2025 Jan 20;4:82. doi: 10.12688/openreseurope.17365.2. eCollection 2024. Open Res Eur. 2025. PMID: 39524113 Free PMC article.
References
-
- Tortonese, E. 1975, Fishes from the Gulf of Aden: Pubblicazioni del Centro di Studio de la Faunistica ed Ecologia Tropicali del CNR: XCIV, Monitore Zoologico Italiano. Supplemento., 6, 167–188.
-
- Bauchot, M.L. and Quignard, J.P.. 1979, Labridae, In: Hureau, J.C. and Monod, T.H. (eds) Check-list of the fishes of the north-eastern Atlantic and of the Mediterranean (CLOFNAM), Paris, France: UNESCO, pp. 426–43.
-
- Oliver, M. and Massuti, M.. 1952, El raó, Xyrichthys novacula (Fam. Labridae) Notas biológicas y biométricas, Bol. Inst. Esp. Oceanogr., 48, 1–15.
-
- Cardinale, M., Colloca, F., and Ardizzone, G.D.. 1997, Feeding ecology of Mediterranean razorfish Xyrichthys novacula in the Tyrrhenian Sea (Central Mediterranean Sea), J. Appl. Ichthyol., 13, 105–111.
-
- Breder, C.M. 1951, Nocturnal and feeding behavior of the labrid fish Xyrichthys psittacus, Copeia, 1951, 162–3.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

