Human APOBEC3B promotes tumor development in vivo including signature mutations and metastases
- PMID: 37797615
- PMCID: PMC10591044
- DOI: 10.1016/j.xcrm.2023.101211
Human APOBEC3B promotes tumor development in vivo including signature mutations and metastases
Abstract
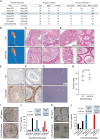
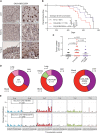
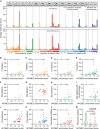
The antiviral DNA cytosine deaminase APOBEC3B has been implicated as a source of mutation in many cancers. However, despite years of work, a causal relationship has yet to be established in vivo. Here, we report a murine model that expresses tumor-like levels of human APOBEC3B. Animals expressing full-body APOBEC3B appear to develop normally. However, adult males manifest infertility, and older animals of both sexes show accelerated rates of carcinogenesis, visual and molecular tumor heterogeneity, and metastasis. Both primary and metastatic tumors exhibit increased frequencies of C-to-T mutations in TC dinucleotide motifs consistent with the established biochemical activity of APOBEC3B. Enrichment for APOBEC3B-attributable single base substitution mutations also associates with elevated levels of insertion-deletion mutations and structural variations. APOBEC3B catalytic activity is required for all of these phenotypes. Together, these studies provide a cause-and-effect demonstration that human APOBEC3B is capable of driving both tumor initiation and evolution in vivo.
Keywords: APOBEC3B; DNA mutagenesis; cancer; lymphoma; murine tumor model; tumor heterogeneity.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





Update of
-
Human APOBEC3B promotes tumor heterogeneity in vivo including signature mutations and metastases.bioRxiv [Preprint]. 2023 Feb 25:2023.02.24.529970. doi: 10.1101/2023.02.24.529970. bioRxiv. 2023. Update in: Cell Rep Med. 2023 Oct 17;4(10):101211. doi: 10.1016/j.xcrm.2023.101211. PMID: 36865194 Free PMC article. Updated. Preprint.
References
-
- Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

