Deciphering D4Z4 CpG methylation gradients in fascioscapulohumeral muscular dystrophy using nanopore sequencing
- PMID: 37798116
- PMCID: PMC10620044
- DOI: 10.1101/gr.277871.123
Deciphering D4Z4 CpG methylation gradients in fascioscapulohumeral muscular dystrophy using nanopore sequencing
Abstract
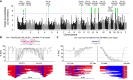
Fascioscapulohumeral muscular dystrophy (FSHD) is caused by a unique genetic mechanism that relies on contraction and hypomethylation of the D4Z4 macrosatellite array on the Chromosome 4q telomere allowing ectopic expression of the DUX4 gene in skeletal muscle. Genetic analysis is difficult because of the large size and repetitive nature of the array, a nearly identical array on the 10q telomere, and the presence of divergent D4Z4 arrays scattered throughout the genome. Here, we combine nanopore long-read sequencing with Cas9-targeted enrichment of 4q and 10q D4Z4 arrays for comprehensive genetic analysis including determination of the length of the 4q and 10q D4Z4 arrays with base-pair resolution. In the same assay, we differentiate 4q from 10q telomeric sequences, determine A/B haplotype, identify paralogous D4Z4 sequences elsewhere in the genome, and estimate methylation for all CpGs in the array. Asymmetric, length-dependent methylation gradients were observed in the 4q and 10q D4Z4 arrays that reach a hypermethylation point at approximately 10 D4Z4 repeat units, consistent with the known threshold of pathogenic D4Z4 contractions. High resolution analysis of individual D4Z4 repeat methylation revealed areas of low methylation near the CTCF/insulator region and areas of high methylation immediately preceding the DUX4 transcriptional start site. Within the DUX4 exons, we observed a waxing/waning methylation pattern with a 180-nucleotide periodicity, consistent with phased nucleosomes. Targeted nanopore sequencing complements recently developed molecular combing and optical mapping approaches to genetic analysis for FSHD by adding precision of the length measurement, base-pair resolution sequencing, and quantitative methylation analysis.
© 2023 Butterfield et al.; Published by Cold Spring Harbor Laboratory Press.
Figures





Update of
-
Deciphering D4Z4 CpG methylation gradients in fascioscapulohumeral muscular dystrophy using nanopore sequencing.bioRxiv [Preprint]. 2023 Mar 29:2023.02.17.528868. doi: 10.1101/2023.02.17.528868. bioRxiv. 2023. Update in: Genome Res. 2023 Sep;33(9):1439-1454. doi: 10.1101/gr.277871.123. PMID: 36824722 Free PMC article. Updated. Preprint.
References
-
- Bouwman LF, den Hamer B, van den Heuvel A, Franken M, Jackson M, Dwyer CA, Tapscott SJ, Rigo F, van der Maarel SM, de Greef JC. 2021. Systemic delivery of a DUX4-targeting antisense oligonucleotide to treat facioscapulohumeral muscular dystrophy. Mol Ther Nucleic Acids 26: 813–827. 10.1016/j.omtn.2021.09.010 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
