Genome-wide association study of placental weight identifies distinct and shared genetic influences between placental and fetal growth
- PMID: 37798380
- PMCID: PMC10632150
- DOI: 10.1038/s41588-023-01520-w
Genome-wide association study of placental weight identifies distinct and shared genetic influences between placental and fetal growth
Abstract
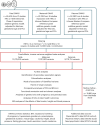
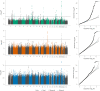
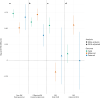
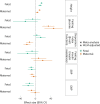
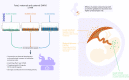
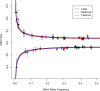
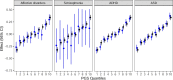
A well-functioning placenta is essential for fetal and maternal health throughout pregnancy. Using placental weight as a proxy for placental growth, we report genome-wide association analyses in the fetal (n = 65,405), maternal (n = 61,228) and paternal (n = 52,392) genomes, yielding 40 independent association signals. Twenty-six signals are classified as fetal, four maternal and three fetal and maternal. A maternal parent-of-origin effect is seen near KCNQ1. Genetic correlation and colocalization analyses reveal overlap with birth weight genetics, but 12 loci are classified as predominantly or only affecting placental weight, with connections to placental development and morphology, and transport of antibodies and amino acids. Mendelian randomization analyses indicate that fetal genetically mediated higher placental weight is causally associated with preeclampsia risk and shorter gestational duration. Moreover, these analyses support the role of fetal insulin in regulating placental weight, providing a key link between fetal and placental growth.
© 2023. The Author(s).
Conflict of interest statement
O.A.A. is a consultant to Cortechs.ai and has received speaker’s fees from Lundbeck, Janssen and Sunovion. B.J.V. is on Allelica’s scientific advisory board. C.J.W.’s spouse works for Regeneron Pharmaceuticals. D.A.L. has received support from Roche Diagnostics and Medtronic for research unrelated to that presented here. H.S.N. has received speaker’s fees from Ferring Pharmaceuticals, Merck A/S, Astra Zeneca, Cook Medical and IBSA Nordic. S.B. has ownerships in Intomics A/S, Hoba Therapeutics Aps, Novo Nordisk A/S, Lundbeck A/S, ALK-Abelló A/S and managing board memberships in Proscion A/S and Intomics A/S. The remaining authors declare no competing interests.
Figures










References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

