Depletion of slow-cycling PDGFRα+ADAM12+ mesenchymal cells promotes antitumor immunity by restricting macrophage efferocytosis
- PMID: 37798557
- PMCID: PMC10602852
- DOI: 10.1038/s41590-023-01642-7
Depletion of slow-cycling PDGFRα+ADAM12+ mesenchymal cells promotes antitumor immunity by restricting macrophage efferocytosis
Abstract
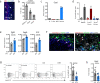
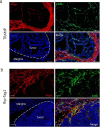
The capacity to survive and thrive in conditions of limited resources and high inflammation is a major driver of tumor malignancy. Here we identified slow-cycling ADAM12+PDGFRα+ mesenchymal stromal cells (MSCs) induced at the tumor margins in mouse models of melanoma, pancreatic cancer and prostate cancer. Using inducible lineage tracing and transcriptomics, we demonstrated that metabolically altered ADAM12+ MSCs induced pathological angiogenesis and immunosuppression by promoting macrophage efferocytosis and polarization through overexpression of genes such as Gas6, Lgals3 and Csf1. Genetic depletion of ADAM12+ cells restored a functional tumor vasculature, reduced hypoxia and acidosis and normalized CAFs, inducing infiltration of effector T cells and growth inhibition of melanomas and pancreatic neuroendocrine cancer, in a process dependent on TGF-β. In human cancer, ADAM12 stratifies patients with high levels of hypoxia and innate resistance mechanisms, as well as factors associated with a poor prognosis and drug resistance such as AXL. Altogether, our data show that depletion of tumor-induced slow-cycling PDGFRα+ MSCs through ADAM12 restores antitumor immunity.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures













References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

