Mitochondrial heteroplasmic shifts reveal a positive selection of breast cancer
- PMID: 37798736
- PMCID: PMC10557196
- DOI: 10.1186/s12967-023-04534-4
Mitochondrial heteroplasmic shifts reveal a positive selection of breast cancer
Abstract
Background: Breast cancer is, despite screening, not always detected early enough and is together with other tumor types known to shed genetic information in circulation. Unlike single-copy nuclear DNA, mitochondrial DNA (mtDNA) copies range from 100s to 10,000s per cell, thus providing a potentially alternative to identify potential missing cancer information in circulation at an early stage.
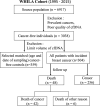
Methods: To characterize mitochondrial mutation landscapes in breast cancer, whole mtDNA sequencing and bioinformatics analyses were performed on 86 breast cancer biopsies and 50 available matched baseline cancer-free whole blood samples from the same individuals, selected from a cohort of middle-aged women in Sweden. To determine whether the mutations can be detected in blood plasma prior to cancer diagnosis, we further designed a nested case-control study (n = 663) and validated the shortlisted mutations using droplet digital PCR.
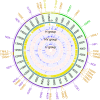
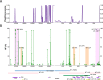
Results: We detected different mutation landscapes between biopsies and matched whole blood samples. Compared to whole blood samples, mtDNA from biopsies had higher heteroplasmic mutations in the D-loop region (P = 0.02), RNR2 (P = 0.005), COX1 (P = 0.037) and CYTB (P = 0.006). Furthermore, the germline mtDNA mutations had higher heteroplasmy level than the lost (P = 0.002) and de novo mutations (P = 0.04). The nonsynonymous to synonymous substitution ratio (dN/dS) was higher for the heteroplasmic mutations (P = 7.25 × 10-12) than that for the homoplasmic mutations, but the de novo (P = 0.06) and lost mutations (P = 0.03) had lower dN/dS than the germline mutations. Interestingly, we found that the critical regions for mitochondrial transcription: MT-HSP1 (odds ratio [OR]: 21.41), MT-TFH (OR: 7.70) and MT-TAS2 (OR: 3.62), had significantly higher heteroplasmic mutations than the rest of the D-loop sub-regions. Finally, we found that the presence of mt.16093T > C mutation increases 67% risk of developing breast cancer.
Conclusions: Our findings show that mitochondrial genetic landscape changes during cancer pathogenesis and positive selection of mtDNA heteroplasmic mutations in breast cancer. Most importantly, the mitochondrial mutations identified in biopsies can be traced back in matched plasma samples and could potentially be used as early breast cancer diagnostic biomarkers.
Keywords: Breast cancer; Heteroplasmic mutation; Sequencing; ddPCR validation; mtDNA.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare that there is no duality of interest associated with this manuscript.
Figures


References
-
- Britt KL, Cuzick J, Phillips KA. Key steps for effective breast cancer prevention. Nat Rev Cancer. 2020;20(8):417–436. - PubMed
-
- Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, et al. Breast cancer. Nat Rev Dis Primers. 2019;5(1):66. - PubMed
-
- Loibl S, Poortmans P, Morrow M, Denkert C, Curigliano G. Breast cancer. Lancet. 2021;397(10286):1750–1769. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

