A DNA origami plasmonic sensor with environment-independent read-out
- PMID: 37799163
- PMCID: PMC10552622
- DOI: 10.1007/s12274-019-2535-0
A DNA origami plasmonic sensor with environment-independent read-out
Abstract
DNA origami is a promising technology for its reproducibility, flexibility, scalability and biocompatibility. Among the several potential applications, DNA origami has been proposed as a tool for drug delivery and as a contrast agent, since a conformational change upon specific target interaction may be used to release a drug or produce a physical signal, respectively. However, its conformation should be robust with respect to the properties of the medium in which either the recognition or the read-out take place, such as pressure, viscosity and any other unspecific interaction other than the desired target recognition. Here we report on the read-out robustness of a tetragonal DNA-origami/gold-nanoparticle hybrid structure able to change its configuration, which is transduced in a change of its plasmonic properties, upon interaction with a specific DNA target. We investigated its response when analyzed in three different media: aqueous solution, solid support and viscous gel. We show that, once a conformational variation is produced, it remains unaffected by the subsequent physical interactions with the environment.
Keywords: DNA origami; gold nanoparticle; molecular detection; plasmonic sensor.
Figures

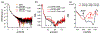
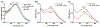

Similar articles
-
Reconfigurable Three-Dimensional Gold Nanorod Plasmonic Nanostructures Organized on DNA Origami Tripod.ACS Nano. 2017 Feb 28;11(2):1172-1179. doi: 10.1021/acsnano.6b06861. Epub 2017 Jan 9. ACS Nano. 2017. PMID: 28056172 Free PMC article.
-
DNA-Nanotechnology-Enabled Chiral Plasmonics: From Static to Dynamic.Acc Chem Res. 2017 Dec 19;50(12):2906-2914. doi: 10.1021/acs.accounts.7b00389. Epub 2017 Sep 27. Acc Chem Res. 2017. PMID: 28953361
-
DNA Origami Guided Self-Assembly of Plasmonic Polymers with Robust Long-Range Plasmonic Resonance.Nano Lett. 2020 Dec 9;20(12):8926-8932. doi: 10.1021/acs.nanolett.0c04055. Epub 2020 Nov 13. Nano Lett. 2020. PMID: 33186046
-
DNA Aptamers for the Functionalisation of DNA Origami Nanostructures.Genes (Basel). 2018 Nov 23;9(12):571. doi: 10.3390/genes9120571. Genes (Basel). 2018. PMID: 30477184 Free PMC article. Review.
-
Dynamic DNA Origami Devices: from Strand-Displacement Reactions to External-Stimuli Responsive Systems.Int J Mol Sci. 2018 Jul 20;19(7):2114. doi: 10.3390/ijms19072114. Int J Mol Sci. 2018. PMID: 30037005 Free PMC article. Review.
References
-
- Ghosh SK; Pal T. Interparticle coupling effect on the surface plasmon resonance of gold nanoparticles: From theory to applications. Chem. Rev 2007, 107, 4797–4862. - PubMed
-
- Reinhard BM; Siu M; Agarwal H; Alivisatos AP; Liphardt J. Calibration of dynamic molecular rulers based on plasmon coupling between gold nanoparticles. Nano Lett. 2005, 5, 2246–2252. - PubMed
-
- Prodan E; Radloff C; Halas NJ; Nordlander P. A hybridization model for the plasmon response of complex nanostructures. Science 2003, 302, 419–422. - PubMed
