Resistance patterns and transmission of mono- and polyresistant TB: clinical impact of WGS
- PMID: 37799267
- PMCID: PMC10549209
- DOI: 10.1093/jacamr/dlad108
Resistance patterns and transmission of mono- and polyresistant TB: clinical impact of WGS
Abstract
Objectives: Rapidly diagnosing drug-resistant TB is crucial for improving treatment and transmission control. WGS is becoming increasingly accessible and has added value to the diagnosis and treatment of TB. The aim of the study was to perform WGS to determine the rate of false-positive results of phenotypic drug susceptibility testing (pDST) and characterize the molecular mechanisms of resistance and transmission of mono- and polyresistant Mycobacterium (M.) tuberculosis.
Methods: WGS was performed on 53 monoresistant and 25 polyresistant M. tuberculosis isolates characterized by pDST. Sequencing data were bioinformatically processed to infer mutations encoding resistance and determine the origin of resistance and phylogenetic relationship between isolates studied.
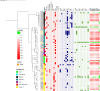
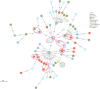
Results: The data showed the variable sensitivity and specificity of WGS in comparison with pDST as the gold standard: isoniazid 92.7% and 92.3%; streptomycin 41.9% and 100.0%; pyrazinamide 15% and 94.8%; and ethambutol 75.0% and 98.6%, respectively. We found novel mutations encoding resistance to streptomycin (in gidB) and pyrazinamide (in kefB). Most isolates belonged to lineage 4 (80.1%) and the overall clustering rate was 11.5%. We observed lineage-specific gene variations encoding resistance to streptomycin and pyrazinamide.
Conclusions: This study highlights the clinical potential of WGS in ruling out false-positive drug resistance following phenotypic or genetic drug testing, and recommend this technology together with the WHO catalogue in designing an optimal individualized treatment regimen and preventing the development of MDR TB. Our results suggest that resistance is primarily developed through spontaneous mutations or selective pressure.
© The Author(s) 2023. Published by Oxford University Press on behalf of British Society for Antimicrobial Chemotherapy.
Figures
References
-
- WHO . Global Tuberculosis Report 2021. 2021. https://www.who.int/publications/i/item/9789240037021.
-
- WHO . Companion Handbook to the WHO Guidelines for the Programmatic Management of Drug-Resistant Tuberculosis. 2014. https://www.ncbi.nlm.nih.gov/books/NBK247420/. - PubMed
